Structure-based discovery of antagonists for GluN3-containing N-methyl-D-aspartate receptors
- PMID: 23973313
- PMCID: PMC3865070
- DOI: 10.1016/j.neuropharm.2013.08.003
Structure-based discovery of antagonists for GluN3-containing N-methyl-D-aspartate receptors
Abstract
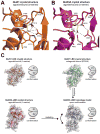
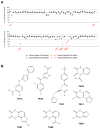
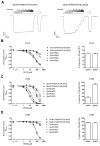
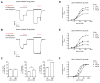
NMDA receptors are ligand-gated ion channels that assemble into tetrameric receptor complexes composed of glycine-binding GluN1 and GluN3 subunits (GluN3A-B) and glutamate-binding GluN2 subunits (GluN2A-D). NMDA receptors can assemble as GluN1/N2 receptors and as GluN3-containing NMDA receptors, which are either glutamate/glycine-activated triheteromeric GluN1/N2/N3 receptors or glycine-activated diheteromeric GluN1/N3 receptors. The glycine-binding GluN1 and GluN3 subunits display strikingly different pharmacological selectivity profiles. However, the pharmacological characterization of GluN3-containing receptors has been hampered by the lack of methods and pharmacological tools to study GluN3 subunit pharmacology in isolation. Here, we have developed a method to study the pharmacology of GluN3 subunits in recombinant diheteromeric GluN1/N3 receptors by mutating the orthosteric ligand-binding pocket in GluN1. This method is suitable for performing compound screening and characterization of structure-activity relationship studies on GluN3 ligands. We have performed a virtual screen of the orthosteric binding site of GluN3A in the search for antagonists with selectivity for GluN3 subunits. In the subsequent pharmacological evaluation of 99 selected compounds, we identified 6-hydroxy-[1,2,5]oxadiazolo[3,4-b]pyrazin-5(4H)-one (TK80) a novel competitive antagonist with preference for the GluN3B subunit. Serendipitously, we also identified [2-hydroxy-5-((4-(pyridin-3-yl)thiazol-2-yl)amino]benzoic acid (TK13) and 4-(2,4-dichlorobenzoyl)-1H-pyrrole-2-carboxylic acid (TK30), two novel non-competitive GluN3 antagonists. These findings demonstrate that structural differences between the orthosteric binding site of GluN3 and GluN1 can be exploited to generate selective ligands.
Keywords: 4-(2,4-dichlorobenzoyl)-1H-pyrrole-2-carboxylic acid; 6-cyano-7-nitroquinoxaline-2,3-dione; 6-hydroxy-[1,2,5]oxadiazolo[3,4-b]pyrazin-5(4H)-one; AMPA; Antagonist; CNQX; DCKA; GluN3 subunit; LBD; N-methyl-d-aspartate; NMDA; NMDA receptor; Selectivity; TEVC; TK13; TK30; TK80; Virtual screening; Xenopus oocyte electrophysiology; [2-hydroxy-5-((4-(pyridin-3-yl)thiazol-2-yl)amino]benzoic acid; dichlorokynurenic acid; iGluR; ionotropic glutamate receptor; ligand-binding domain; two-electrode voltage-clamp; α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Negative allosteric modulation of GluN1/GluN3 NMDA receptors.Neuropharmacology. 2020 Oct 1;176:108117. doi: 10.1016/j.neuropharm.2020.108117. Epub 2020 May 7. Neuropharmacology. 2020. PMID: 32389749 Free PMC article.
-
Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.J Biol Chem. 2013 Nov 15;288(46):33124-35. doi: 10.1074/jbc.M113.480210. Epub 2013 Sep 26. J Biol Chem. 2013. PMID: 24072709 Free PMC article.
-
Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.Proc Natl Acad Sci U S A. 2017 Aug 15;114(33):E6942-E6951. doi: 10.1073/pnas.1707752114. Epub 2017 Jul 31. Proc Natl Acad Sci U S A. 2017. PMID: 28760974 Free PMC article.
-
Glycine agonism in ionotropic glutamate receptors.Neuropharmacology. 2021 Aug 1;193:108631. doi: 10.1016/j.neuropharm.2021.108631. Epub 2021 May 28. Neuropharmacology. 2021. PMID: 34058193 Review.
-
The GluN3-containing NMDA receptors.Channels (Austin). 2025 Dec;19(1):2490308. doi: 10.1080/19336950.2025.2490308. Epub 2025 Apr 16. Channels (Austin). 2025. PMID: 40235311 Free PMC article. Review.
Cited by
-
Allosteric modulation of GluN1/GluN3 NMDA receptors by GluN1-selective competitive antagonists.J Gen Physiol. 2023 Jun 5;155(6):e202313340. doi: 10.1085/jgp.202313340. Epub 2023 Apr 20. J Gen Physiol. 2023. PMID: 37078900 Free PMC article.
-
Plasticity in the Functional Properties of NMDA Receptors Improves Network Stability during Severe Energy Stress.J Neurosci. 2024 Feb 28;44(9):e0502232024. doi: 10.1523/JNEUROSCI.0502-23.2024. J Neurosci. 2024. PMID: 38262722 Free PMC article.
-
Structural features in the glycine-binding sites of the GluN1 and GluN3A subunits regulate the surface delivery of NMDA receptors.Sci Rep. 2019 Aug 23;9(1):12303. doi: 10.1038/s41598-019-48845-3. Sci Rep. 2019. PMID: 31444392 Free PMC article.
-
Activation of excitatory glycine NMDA receptors: At the mercy of a whimsical GluN1 subunit.J Gen Physiol. 2023 Jun 5;155(6):e202313391. doi: 10.1085/jgp.202313391. Epub 2023 May 3. J Gen Physiol. 2023. PMID: 37133818 Free PMC article.
-
Derivatives of (R)-3-(5-Furanyl)carboxamido-2-aminopropanoic Acid as Potent NMDA Receptor Glycine Site Agonists with GluN2 Subunit-Specific Activity.J Med Chem. 2022 Jan 13;65(1):734-746. doi: 10.1021/acs.jmedchem.1c01810. Epub 2021 Dec 17. J Med Chem. 2022. PMID: 34918931 Free PMC article.
References
-
- Andersson O, Stenqvist A, Attersand A, von Euler G. Nucleotide sequence, genomic organization, and chromosomal localization of genes encoding the human NMDA receptor subunits NR3A and NR3B. Genomics. 2001;78:178–184. - PubMed
-
- Awobuluyi M, Yang J, Ye Y, Chatterton JE, Godzik A, Lipton SA, Zhang D. Subunit-specific roles of glycine-binding domains in activation of NR1/NR3 N-methyl-D-aspartate receptors. Mol Pharmacol. 2007;71:112–122. - PubMed
-
- Bjerrum EJ, Biggin PC. Rigid body essential X-ray crystallography: distinguishing the bend and twist of glutamate receptor ligand binding domains. Proteins. 2008;72:434–446. - PubMed
-
- Cavara NA, Hollmann M. Shuffling the deck anew: how NR3 tweaks NMDA receptor function. Mol Neurobiol. 2008;38:16–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

