Novel feature of Mycobacterium avium subsp. paratuberculosis, highlighted by characterization of the heparin-binding hemagglutinin adhesin
- PMID: 23974028
- PMCID: PMC3807500
- DOI: 10.1128/JB.00671-13
Novel feature of Mycobacterium avium subsp. paratuberculosis, highlighted by characterization of the heparin-binding hemagglutinin adhesin
Abstract
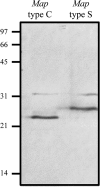
Mycobacterium avium subsp. paratuberculosis comprises two genotypically defined groups, known as the cattle (C) and sheep (S) groups. Recent studies have reported phenotypic differences between M. avium subsp. paratuberculosis groups C and S, including growth rates, infectivity for macrophages, and iron metabolism. In this study, we investigated the genotypes and biological properties of the virulence factor heparin-binding hemagglutinin adhesin (HBHA) for both groups. In Mycobacterium tuberculosis, HBHA is a major adhesin involved in mycobacterium-host interactions and extrapulmonary dissemination of infection. To investigate HBHA in M. avium subsp. paratuberculosis, we studied hbhA polymorphisms by fragment analysis using the GeneMapper technology across a large collection of isolates genotyped by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) and IS900 restriction fragment length polymorphism (RFLP-IS900) analyses. Furthermore, we analyzed the structure-function relationships of recombinant HBHA proteins of types C and S by heparin-Sepharose chromatography and surface plasmon resonance (SPR) analyses. In silico analysis revealed two forms of HBHA, corresponding to the prototype genomes for the C and S types of M. avium subsp. paratuberculosis. This observation was confirmed using GeneMapper on 85 M. avium subsp. paratuberculosis strains, including 67 strains of type C and 18 strains of type S. We found that HBHAs from all type C strains contain a short C-terminal domain, while those of type S present a long C-terminal domain, similar to that produced by Mycobacterium avium subsp. avium. The purification of recombinant HBHA from M. avium subsp. paratuberculosis of both types by heparin-Sepharose chromatography highlighted a correlation between their affinities for heparin and the lengths of their C-terminal domains, which was confirmed by SPR analysis. Thus, types C and S of M. avium subsp. paratuberculosis may be distinguished by the types of HBHA they produce, which differ in size and adherence properties, thereby providing new evidence that strengthens the genotypic differences between the C and S types of M. avium subsp. paratuberculosis.
Figures





References
-
- Biet F, Sevilla IA, Cochard T, Lefrancois LH, Garrido JM, Heron I, Juste RA, McLuckie J, Thibault VC, Supply P, Collins DM, Behr MA, Stevenson K. 2012. Inter- and intra-subtype genotypic differences that differentiate Mycobacterium avium subspecies paratuberculosis strains. BMC Microbiol. 12:264. 10.1186/1471-2180-12-264 - DOI - PMC - PubMed
-
- Castellanos E, Romero B, Rodriguez S, de Juan L, Bezos J, Mateos A, Dominguez L, Aranaz A. 2010. Molecular characterization of Mycobacterium avium subspecies paratuberculosis types II and III isolates by a combination of MIRU-VNTR loci. Vet. Microbiol. 144:118–126 - PubMed
-
- Castellanos E, Aranaz A, Gould KA, Linedale R, Stevenson K, Alvarez J, Dominguez L, de Juan L, Hinds J, Bull TJ. 2009. Discovery of stable and variable differences in the Mycobacterium avium subsp. paratuberculosis type I, II, and III genomes by pan-genome microarray analysis. Appl. Environ. Microbiol. 75:676–686 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

