Killer cell immunoglobulin-like receptor (KIR) genes and their HLA-C ligands in a Ugandan population
- PMID: 23974321
- PMCID: PMC3824577
- DOI: 10.1007/s00251-013-0724-7
Killer cell immunoglobulin-like receptor (KIR) genes and their HLA-C ligands in a Ugandan population
Abstract
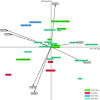
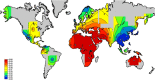
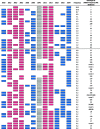
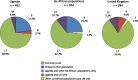
Killer cell immunoglobulin-like receptor (KIR) genes are expressed by natural killer cells and encoded by a family of genes exhibiting considerable haplotypic and allelic variation. HLA-C molecules, the dominant ligands for KIR, are present in all individuals and are discriminated by two KIR epitopes, C1 and C2. We studied the frequencies of KIR genes and HLA-C1 and C2 groups in a large cohort (n = 492) from Kampala, Uganda, East Africa and compared our findings with published data from other populations in sub-Saharan Africa (SSA) and several European populations. We find considerably more KIR diversity and weaker linkage disequilibrium in SSA compared to the European populations and describe several novel KIR genotypes. C1 and C2 frequencies were similar to other SSA populations with a higher frequency of the C2 epitope (54.9 %) compared to Europe (average 39.7 %). Analysis of this large cohort from Uganda in the context of other African populations reveals variations in KIR and HLA-C1 and C2 that are consistent with migrations within Africa and potential selection pressures on these genes. Our results will help understand how KIR/HLA-C interactions contribute to resistance to pathogens and reproductive success.
Figures

Similar articles
-
Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans.PLoS Genet. 2013 Oct;9(10):e1003938. doi: 10.1371/journal.pgen.1003938. Epub 2013 Oct 31. PLoS Genet. 2013. PMID: 24204327 Free PMC article.
-
Killer-cell immunoglobulin-like receptor (KIR) and human leukocyte antigen (HLA) class I genetic diversity in four South African populations.Hum Immunol. 2017 Jul-Aug;78(7-8):503-509. doi: 10.1016/j.humimm.2017.05.006. Epub 2017 May 29. Hum Immunol. 2017. PMID: 28571758
-
Diversity of KIR, HLA Class I, and Their Interactions in Seven Populations of Sub-Saharan Africans.J Immunol. 2019 May 1;202(9):2636-2647. doi: 10.4049/jimmunol.1801586. Epub 2019 Mar 27. J Immunol. 2019. PMID: 30918042 Free PMC article.
-
KIR specificity and avidity of standard and unusual C1, C2, Bw4, Bw6 and A3/11 amino acid motifs at entire HLA:KIR interface between NK and target cells, the functional and evolutionary classification of HLA class I molecules.Int J Immunogenet. 2019 Aug;46(4):217-231. doi: 10.1111/iji.12433. Epub 2019 Jun 18. Int J Immunogenet. 2019. PMID: 31210416 Review.
-
Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules.Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):800-11. doi: 10.1098/rstb.2011.0266. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312047 Free PMC article. Review.
Cited by
-
The KIR repertoire of a West African chimpanzee population is characterized by limited gene, allele, and haplotype variation.Front Immunol. 2023 Dec 11;14:1308316. doi: 10.3389/fimmu.2023.1308316. eCollection 2023. Front Immunol. 2023. PMID: 38149259 Free PMC article.
-
Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes.J Immunol. 2015 Oct 1;195(7):3160-70. doi: 10.4049/jimmunol.1501358. Epub 2015 Aug 26. J Immunol. 2015. PMID: 26311903 Free PMC article.
-
Association of Inhibitory Killer Cell Immunoglobulin-like Receptor Ligands With Higher Plasmodium falciparum Parasite Prevalence.J Infect Dis. 2021 Jul 2;224(1):175-183. doi: 10.1093/infdis/jiaa698. J Infect Dis. 2021. PMID: 33165540 Free PMC article.
-
High KIR diversity in Uganda and Botswana children living with HIV.bioRxiv [Preprint]. 2024 Dec 7:2024.12.03.626612. doi: 10.1101/2024.12.03.626612. bioRxiv. 2024. PMID: 39677597 Free PMC article. Preprint.
-
Immunology in Africa.Trop Med Int Health. 2015 Dec;20(12):1771-7. doi: 10.1111/tmi.12599. Epub 2015 Oct 12. Trop Med Int Health. 2015. PMID: 26391634 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

