DeNovoGear: de novo indel and point mutation discovery and phasing
- PMID: 23975140
- PMCID: PMC4003501
- DOI: 10.1038/nmeth.2611
DeNovoGear: de novo indel and point mutation discovery and phasing
Abstract
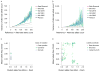
We present DeNovoGear software for analyzing de novo mutations from familial and somatic tissue sequencing data. DeNovoGear uses likelihood-based error modeling to reduce the false positive rate of mutation discovery in exome analysis and fragment information to identify the parental origin of germ-line mutations. We used DeNovoGear on human whole-genome sequencing data to produce a set of predicted de novo insertion and/or deletion (indel) mutations with a 95% validation rate.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

