Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males
- PMID: 23975186
- PMCID: PMC3883776
- DOI: 10.4161/epi.25798
Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males
Abstract
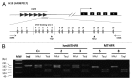
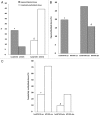
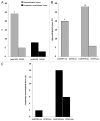
Aberrant methylation at the H19 paternal imprinted gene has been identified in different cohorts of infertile males. The causes of H19 methylation errors are poorly understood. In this study, we investigated the methylation status of the H19 gene in semen DNA samples from infertile males affected by MTHFR gene promoter hypermethylation. DNA from normal and abnormal semen samples harbouring MTHFR gene promoter hypermethylated, hmMTHFR-nor and hmMTHFR-abn, and without MTHFR methylation, MTHFR-nor and MTHFR-abn, were investigated for methylation status in the H19 locus using bisulfite-treated DNA PCR, followed by cloning and sequencing. The prevalence of H19 hypomethylated clones was 20% in hmMTHFR-nor and 0% in MTHFR-nor semen samples (p<0.05), and 28% in hmMTHFR-abn compared with 16% in MTHFR-abn semen samples (p>0.05). These results underscore the association between H19 methylation defects and hypermethylation of the MTHFR gene promoter in normal semen samples and suggest that aberrant methylation at H19 may occur in the normal sperm of infertile males affected by MTHFR gene dysfunction. These findings provide new insights into the mechanisms causing abnormal methylation in imprinted genes and, in turn, male infertility.
Keywords: DNA methylation; genetic imprinting; male infertility; spermatozoa.
Figures

References
-
- Diplas AI, Lambertini L, Lee MJ, Sperling R, Lee YL, Wetmur J, et al. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4:235–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
