Quantitative Proteomics Identifies Activation of Hallmark Pathways of Cancer in Patient Melanoma
- PMID: 23976835
- PMCID: PMC3748992
- DOI: 10.4172/jpb.1000260
Quantitative Proteomics Identifies Activation of Hallmark Pathways of Cancer in Patient Melanoma
Abstract
Molecular pathways regulating melanoma initiation and progression are potential targets of therapeutic development for this aggressive cancer. Identification and molecular analysis of these pathways in patients has been primarily restricted to targeted studies on individual proteins. Here, we report the most comprehensive analysis of formalin-fixed paraffin-embedded human melanoma tissues using quantitative proteomics. From 61 patient samples, we identified 171 proteins varying in abundance among benign nevi, primary melanoma, and metastatic melanoma. Seventy-three percent of these proteins were validated by immunohistochemistry staining of malignant melanoma tissues from the Human Protein Atlas database. Our results reveal that molecular pathways involved with tumor cell proliferation, motility, and apoptosis are mis-regulated in melanoma. These data provide the most comprehensive proteome resource on patient melanoma and reveal insight into the molecular mechanisms driving melanoma progression.
Keywords: FFPE tissue; Mass spectrometry; Melanoma; Proteomics; Quantitative.
Figures

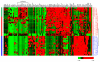
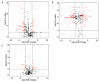


References
-
- Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, et al. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–6243. - PubMed
-
- Markovic SN, Erickson LA, Rao RD, Weenig RH, Pockaj BA, et al. Malignant melanoma in the 21st century, part 1: epidemiology, risk factors, screening, prevention, and diagnosis. Mayo Clin Proc. 2007;82:364–380. - PubMed
-
- ACS . Cancer Facts and Figures 2012. American Cancer Society; Atlanta: 2012.
-
- Helmbach H, Rossmann E, Kern MA, Schadendorf D. Drug-resistance in human melanoma. Int J Cancer. 2001;93:617–622. - PubMed
-
- Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–3151. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
