The HILDA complex coordinates a conditional switch in the 3'-untranslated region of the VEGFA mRNA
- PMID: 23976881
- PMCID: PMC3747992
- DOI: 10.1371/journal.pbio.1001635
The HILDA complex coordinates a conditional switch in the 3'-untranslated region of the VEGFA mRNA
Abstract
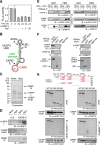
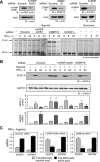
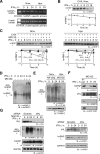
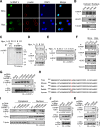
Cell regulatory circuits integrate diverse, and sometimes conflicting, environmental cues to generate appropriate, condition-dependent responses. Here, we elucidate the components and mechanisms driving a protein-directed RNA switch in the 3'UTR of vascular endothelial growth factor (VEGF)-A. We describe a novel HILDA (hypoxia-inducible hnRNP L-DRBP76-hnRNP A2/B1) complex that coordinates a three-element RNA switch, enabling VEGFA mRNA translation during combined hypoxia and inflammation. In addition to binding the CA-rich element (CARE), heterogeneous nuclear ribonucleoprotein (hnRNP) L regulates switch assembly and function. hnRNP L undergoes two previously unrecognized, condition-dependent posttranslational modifications: IFN-γ induces prolyl hydroxylation and von Hippel-Lindau (VHL)-mediated proteasomal degradation, whereas hypoxia stimulates hnRNP L phosphorylation at Tyr(359), inducing binding to hnRNP A2/B1, which stabilizes the protein. Also, phospho-hnRNP L recruits DRBP76 (double-stranded RNA binding protein 76) to the 3'UTR, where it binds an adjacent AU-rich stem-loop (AUSL) element, "flipping" the RNA switch by disrupting the GAIT (interferon-gamma-activated inhibitor of translation) element, preventing GAIT complex binding, and driving robust VEGFA mRNA translation. The signal-dependent, HILDA complex coordinates the function of a trio of neighboring RNA elements, thereby regulating translation of VEGFA and potentially other mRNA targets. The VEGFA RNA switch might function to ensure appropriate angiogenesis and tissue oxygenation during conflicting signals from combined inflammation and hypoxia. We propose the VEGFA RNA switch as an archetype for signal-activated, protein-directed, multi-element RNA switches that regulate posttranscriptional gene expression in complex environments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Tammela T, Enholm B, Alitalo K, Paavonen K (2005) The biology of vascular endothelial growth factors. Cardiovasc Res 65: 550–563. - PubMed
-
- Hua Z, Lv Q, Ye W, Wong CK, Cai G, et al. (2006) MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1: e116 doi:10.1371/journal.pone.0000116 - DOI - PMC - PubMed
-
- Pages G, Pouyssegur J (2005) Transcriptional regulation of the Vascular Endothelial Growth Factor gene—a concert of activating factors. Cardiovasc Res 65: 564–573. - PubMed
-
- Ferrara N, Davis-Smyth T (1997) The biology of vascular endothelial growth factor. Endocr Rev 18: 4–25. - PubMed
-
- Weis SM, Cheresh DA (2005) Pathophysiological consequences of VEGF-induced vascular permeability. Nature 437: 497–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

