Raman spectroscopic imaging of the whole Ciona intestinalis embryo during development
- PMID: 23977129
- PMCID: PMC3748111
- DOI: 10.1371/journal.pone.0071739
Raman spectroscopic imaging of the whole Ciona intestinalis embryo during development
Abstract
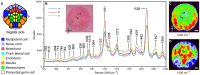
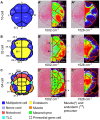
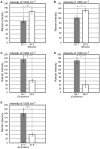
Intracellular composition and the distribution of bio-molecules play central roles in the specification of cell fates and morphogenesis during embryogenesis. Consequently, investigation of changes in the expression and distribution of bio-molecules, especially mRNAs and proteins, is an important challenge in developmental biology. Raman spectroscopic imaging, a non-invasive and label-free technique, allows simultaneous imaging of the intracellular composition and distribution of multiple bio-molecules. In this study, we explored the application of Raman spectroscopic imaging in the whole Ciona intestinalis embryo during development. Analysis of Raman spectra scattered from C. intestinalis embryos revealed a number of localized patterns of high Raman intensity within the embryo. Based on the observed distribution of bio-molecules, we succeeded in identifying the location and structure of differentiated muscle and endoderm within the whole embryo, up to the tailbud stage, in a label-free manner. Furthermore, during cell differentiation, we detected significant differences in cell state between muscle/endoderm daughter cells and daughter cells with other fates that had divided from the same mother cells; this was achieved by focusing on the Raman intensity of single Raman bands at 1002 or 1526 cm(-1), respectively. This study reports the first application of Raman spectroscopic imaging to the study of identifying and characterizing differentiating tissues in a whole chordate embryo. Our results suggest that Raman spectroscopic imaging is a feasible label-free technique for investigating the developmental process of the whole embryo of C. intestinalis.
Conflict of interest statement
Figures

References
-
- Das RS, Agrawal YK (2011) Raman spectroscopy: Recent advancements, techniques and applications. Vibrational Spectroscopy 57: 163–176.
-
- McNay G, Eustace D, Smith WE, Faulds K, Graham D (2011) Surface-enhanced Raman scattering (SERS) and surface-enhanced resonance Raman scattering (SERRS): a review of applications. Applied spectroscopy 65: 825–837. - PubMed
-
- Krafft C, Knetschke T, Funk RHW, Salzer R (2006) Studies on stress-induced changes at the subcellular level by Raman microspectroscopic mapping. Analytical chemistry 78: 4424–4429. - PubMed
-
- Huang C-K, Ando M, Hamaguchi H, Shigeto S (2012) Disentangling dynamic changes of multiple cellular components during the yeast cell cycle by in vivo multivariate Raman imaging. Analytical chemistry 84: 5661–5668. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

