Size of gene specific inverted repeat--dependent gene deletion In Saccharomyces cerevisiae
- PMID: 23977230
- PMCID: PMC3748122
- DOI: 10.1371/journal.pone.0072137
Size of gene specific inverted repeat--dependent gene deletion In Saccharomyces cerevisiae
Abstract
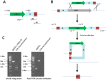
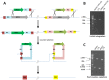
We describe here an approach for rapidly producing scar-free and precise gene deletions in S. cerevisiae with high efficiency. Preparation of the disruption gene cassette in this approach was simply performed by overlap extension-PCR of an invert repeat of a partial or complete sequence of the targeted gene with URA3. Integration of the prepared disruption gene cassette to the designated position of a target gene leads to the formation of a mutagenesis cassette within the yeast genome, which consists of a URA3 gene flanked by the targeted gene and its inverted repeat between two short identical direct repeats. The inherent instability of the inverted sequences in close proximity facilitates the self-excision of the entire mutagenesis cassette deposited in the genome and promotes homologous recombination resulting in a seamless deletion via a single transformation. This rapid assembly circumvents the difficulty during preparation of disruption gene cassettes composed of two inverted repeats of the URA3, which requires the engineering of unique restriction sites for subsequent digestion and T4 DNA ligation in vitro. We further identified that the excision of the entire mutagenesis cassette flanked by two DRs in the transformed S. cerevisiae is dependent on the length of the inverted repeat of which a minimum of 800 bp is required for effective gene deletion. The deletion efficiency improves with the increase of the inverted repeat till 1.2 kb. Finally, the use of gene-specific inverted repeats of target genes enables simultaneous gene deletions. The procedure has the potential for application on other yeast strains to achieve precise and efficient removal of gene sequences.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

