Analysis of human blood plasma proteome from ten healthy volunteers from Indian population
- PMID: 23977322
- PMCID: PMC3748081
- DOI: 10.1371/journal.pone.0072584
Analysis of human blood plasma proteome from ten healthy volunteers from Indian population
Abstract
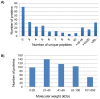
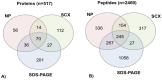
Analysis of any mammalian plasma proteome is a challenge, particularly by mass spectrometry, due to the presence of albumin and other abundant proteins which can mask the detection of low abundant proteins. As detection of human plasma proteins is valuable in diagnostics, exploring various workflows with minimal fractionation prior to mass spectral analysis, is required in order to study population diversity involving analysis in a large cohort of samples. Here, we used 'reference plasma sample', a pool of plasma from 10 healthy individuals from Indian population in the age group of 25-60 yrs including 5 males and 5 females. The 14 abundant proteins were immunodepleted from plasma and then evaluated by three different workflows for proteome analysis using a nanoflow reverse phase liquid chromatography system coupled to a LTQ Orbitrap Velos mass spectrometer. The analysis of reference plasma sample a) without prefractionation, b) after prefractionation at peptide level by strong cation exchange chromatography and c) after prefractionation at protein level by sodium dodecyl sulfate polyacrylamide gel electrophoresis, led to the identification of 194, 251 and 342 proteins respectively. Together, a comprehensive dataset of 517 unique proteins was achieved from all the three workflows, including 271 proteins with high confidence identified by ≥ 2 unique peptides in any of the workflows or identified by single peptide in any of the two workflows. A total of 70 proteins were common in all the three workflows. Some of the proteins were unique to our study and could be specific to Indian population. The high-confidence dataset obtained from our study may be useful for studying the population diversity, in discovery and validation process for biomarker identification.
Conflict of interest statement
Figures



References
-
- Anderson NL, Anderson NG (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1: 845–867. - PubMed
-
- Hortin GL, Sviridov D, Anderson NL (2008) High-abundance polypeptides of the human plasma proteome comprising the top 4 logs of polypeptide abundance. Clin Chem 4: 1608–1616. - PubMed
-
- Anderson NL (2010) The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem 56: 177–185. - PubMed
-
- Surinova S, Schiess R, Hüttenhain R, Cerciello F, Wollscheid B, et al. (2011) On the development of plasma protein biomarkers. J Proteome Res 10: 5–16. - PubMed
-
- Altelaar AFM, Heck AJR (2011) Trends in ultrasensitive proteomics. Curr Op Chem Biol 16: 1–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

