A new approach to measuring protein backbone protection with high spatial resolution using H/D exchange and electron capture dissociation
- PMID: 23978257
- PMCID: PMC3801164
- DOI: 10.1021/ac401868b
A new approach to measuring protein backbone protection with high spatial resolution using H/D exchange and electron capture dissociation
Abstract
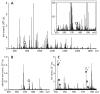
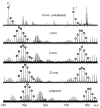
Inadequate spatial resolution remains one of the most serious limitations of hydrogen/deuterium exchange-mass spectrometry (HDX-MS), especially when applied to larger proteins (over 30 kDa). Supplementing proteolytic fragmentation of the protein in solution with ion dissociation in the gas phase has been used successfully by several groups to obtain near-residue level resolution. However, the restrictions imposed by the LC-MS/MS mode of operation on the data acquisition time frame makes it difficult in many cases to obtain a signal-to-noise ratio adequate for reliable assignment of the backbone amide protection levels at individual residues. This restriction is lifted in the present work by eliminating the LC separation step from the workflow and taking advantage of the high resolving power and dynamic range of a Fourier transform ion cyclotron resonance-mass spectrometer (FTICR-MS). A residue-level resolution is demonstrated for a peptic fragment of a 37 kDa recombinant protein (N-lobe of human serum transferrin), using electron-capture dissociation as an ion fragmentation tool. The absence of hydrogen scrambling in the gas phase prior to ion dissociation is verified using redundant HDX-MS data generated by FTICR-MS. The backbone protection pattern generated by direct HDX-MS/MS is in excellent agreement with the known crystal structure of the protein but also provides information on conformational dynamics, which is not available from the static X-ray structure.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

