Statistical mechanical treatments of protein amyloid formation
- PMID: 23979423
- PMCID: PMC3794734
- DOI: 10.3390/ijms140917420
Statistical mechanical treatments of protein amyloid formation
Abstract
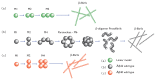
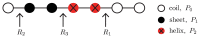
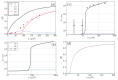
Protein aggregation is an important field of investigation because it is closely related to the problem of neurodegenerative diseases, to the development of biomaterials, and to the growth of cellular structures such as cyto-skeleton. Self-aggregation of protein amyloids, for example, is a complicated process involving many species and levels of structures. This complexity, however, can be dealt with using statistical mechanical tools, such as free energies, partition functions, and transfer matrices. In this article, we review general strategies for studying protein aggregation using statistical mechanical approaches and show that canonical and grand canonical ensembles can be used in such approaches. The grand canonical approach is particularly convenient since competing pathways of assembly and dis-assembly can be considered simultaneously. Another advantage of using statistical mechanics is that numerically exact solutions can be obtained for all of the thermodynamic properties of fibrils, such as the amount of fibrils formed, as a function of initial protein concentration. Furthermore, statistical mechanics models can be used to fit experimental data when they are available for comparison.
Figures









Similar articles
-
A statistical mechanical approach to protein aggregation.J Chem Phys. 2011 Dec 21;135(23):235102. doi: 10.1063/1.3666837. J Chem Phys. 2011. PMID: 22191902
-
Mechanical properties of amyloid-like fibrils defined by secondary structures.Nanoscale. 2015 May 7;7(17):7745-52. doi: 10.1039/c4nr05109b. Nanoscale. 2015. PMID: 25839069
-
Self-folding and aggregation of amyloid nanofibrils.Nanoscale. 2011 Apr;3(4):1748-55. doi: 10.1039/c0nr00840k. Epub 2011 Feb 23. Nanoscale. 2011. PMID: 21347488
-
Protein Co-Aggregation Related to Amyloids: Methods of Investigation, Diversity, and Classification.Int J Mol Sci. 2018 Aug 4;19(8):2292. doi: 10.3390/ijms19082292. Int J Mol Sci. 2018. PMID: 30081572 Free PMC article. Review.
-
Protein denaturation and aggregation: Cellular responses to denatured and aggregated proteins.Ann N Y Acad Sci. 2005 Dec;1066:181-221. doi: 10.1196/annals.1363.030. Ann N Y Acad Sci. 2005. PMID: 16533927 Review.
References
-
- Thirumalai D., Klimov D., Dima R. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr. Opin. Struct. Biol. 2003;13:146–159. - PubMed
-
- Ma B., Nussinov R. Simulations as analytical tools to understand protein aggregation and predict amyloid conformation. Curr. Opin. Chem. Biol. 2006;10:445–452. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

