Multipoint binding of the SLP-76 SH2 domain to ADAP is critical for oligomerization of SLP-76 signaling complexes in stimulated T cells
- PMID: 23979596
- PMCID: PMC3811887
- DOI: 10.1128/MCB.00410-13
Multipoint binding of the SLP-76 SH2 domain to ADAP is critical for oligomerization of SLP-76 signaling complexes in stimulated T cells
Abstract
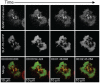
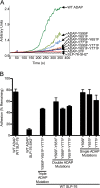
The adapter molecules SLP-76 and LAT play central roles in T cell activation by recruiting enzymes and other adapters into multiprotein complexes that coordinate highly regulated signal transduction pathways. While many of the associated proteins have been characterized, less is known concerning the mechanisms of assembly for these dynamic and potentially heterogeneous signaling complexes. Following T cell receptor (TCR) stimulation, SLP-76 is found in structures called microclusters, which contain many signaling complexes. Previous studies showed that a mutation to the SLP-76 C-terminal SH2 domain nearly abolished SLP-76 microclusters, suggesting that the SH2 domain facilitates incorporation of signaling complexes into microclusters. S. C. Bunnell, A. L. Singer, D. I. Hong, B. H. Jacque, M. S. Jordan, M. C. Seminario, V. A. Barr, G. A. Koretzky, and L. E. Samelson, Mol. Cell. Biol., 26:7155-7166, 2006). Using biophysical methods, we demonstrate that the adapter, ADAP, contains three binding sites for SLP-76, and that multipoint binding to ADAP fragments oligomerizes the SLP-76 SH2 domain in vitro. These results were complemented with confocal imaging and functional studies of cells expressing ADAP with various mutations. Our results demonstrate that all three binding sites are critical for SLP-76 microcluster assembly, but any combination of two sites will partially induce microclusters. These data support a model whereby multipoint binding of SLP-76 to ADAP facilitates the assembly of SLP-76 microclusters. This model has implications for the regulation of SLP-76 and LAT microclusters and, as a result, T cell signaling.
Figures





References
-
- Sangani D, Venien-Bryan C, Harder T. 2009. Phosphotyrosine-dependent in vitro reconstitution of recombinant LAT-nucleated multiprotein signalling complexes on liposomes. Mol. Membr. Biol. 26:159–170 - PubMed
-
- Houtman JC, Higashimoto Y, Dimasi N, Cho S, Yamaguchi H, Bowden B, Regan C, Malchiodi EL, Mariuzza R, Schuck P, Appella E, Samelson LE. 2004. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry 43:4170–4178 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
