Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment
- PMID: 23979677
- PMCID: PMC3903129
- DOI: 10.1038/ncomms3120
Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment
Abstract
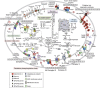
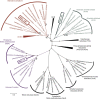
Microorganisms in the subsurface represent a substantial but poorly understood component of the Earth's biosphere. Subsurface environments are complex and difficult to characterize; thus, their microbiota have remained as a 'dark matter' of the carbon and other biogeochemical cycles. Here we deeply sequence two sediment-hosted microbial communities from an aquifer adjacent to the Colorado River, CO, USA. No single organism represents more than ~1% of either community. Remarkably, many bacteria and archaea in these communities are novel at the phylum level or belong to phyla lacking a sequenced representative. The dominant organism in deeper sediment, RBG-1, is a member of a new phylum. On the basis of its reconstructed complete genome, RBG-1 is metabolically versatile. Its wide respiration-based repertoire may enable it to respond to the fluctuating redox environment close to the water table. We document extraordinary microbial novelty and the importance of previously unknown lineages in sediment biogeochemical transformations.
Figures


References
-
- Lipp J. S., Morono Y., Inagaki F. & Hinrichs K. U. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994 (2008). - PubMed
-
- Archer D. Methane hydrate stability and anthropogenic climate change. Biogeosciences 4, 521–544 (2007).
-
- Wadham J. L. et al. Potential methane reservoirs beneath Antarctica. Nature 488, 633–637 (2012). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

