Identification of transcription factor binding sites from ChIP-seq data at high resolution
- PMID: 23980024
- PMCID: PMC3799470
- DOI: 10.1093/bioinformatics/btt470
Identification of transcription factor binding sites from ChIP-seq data at high resolution
Abstract
Motivation: Chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-seq) is widely used to study the in vivo binding sites of transcription factors (TFs) and their regulatory targets. Recent improvements to ChIP-seq, such as increased resolution, promise deeper insights into transcriptional regulation, yet require novel computational tools to fully leverage their advantages.
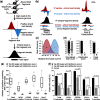
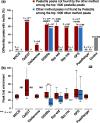
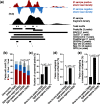
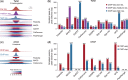
Results: To this aim, we have developed peakzilla, which can identify closely spaced TF binding sites at high resolution (i.e. resolves individual binding sites even if spaced closely), as we demonstrate using semisynthetic datasets, performing ChIP-seq for the TF Twist in Drosophila embryos with different experimental fragment sizes, and analyzing ChIP-exo datasets. We show that the increased resolution reached by peakzilla is highly relevant, as closely spaced Twist binding sites are strongly enriched in transcriptional enhancers, suggesting a signature to discriminate functional from abundant non-functional or neutral TF binding. Peakzilla is easy to use, as it estimates all the necessary parameters from the data and is freely available.
Availability and implementation: The peakzilla program is available from https://github.com/steinmann/peakzilla or http://www.starklab.org/data/peakzilla/.
Contact: stark@starklab.org.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

References
-
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. - PubMed
-
- Bardet AF, et al. A computational pipeline for comparative ChIP-seq analyses. Nat. Protoc. 2012;7:45–61. - PubMed
-
- Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

