Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity
- PMID: 23980144
- PMCID: PMC3773741
- DOI: 10.1073/pnas.1310894110
Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity
Abstract
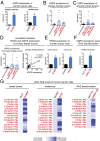
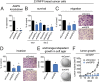
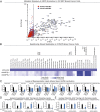
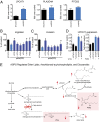
Aberrant lipid metabolism is an established hallmark of cancer cells. In particular, ether lipid levels have been shown to be elevated in tumors, but their specific function in cancer remains elusive. We show here that the metabolic enzyme alkylglyceronephosphate synthase (AGPS), a critical step in the synthesis of ether lipids, is up-regulated across multiple types of aggressive human cancer cells and primary tumors. We demonstrate that ablation of AGPS in cancer cells results in reduced cell survival, cancer aggressiveness, and tumor growth through altering the balance of ether lipid, fatty acid, eicosanoid, and fatty acid-derived glycerophospholipid metabolism, resulting in an overall reduction in the levels of several oncogenic signaling lipids. Taken together, our results reveal that AGPS, in addition to maintaining ether lipids, also controls cellular utilization of fatty acids, favoring the generation of signaling lipids necessary for promoting the aggressive features of cancer.
Keywords: cancer metabolism; eicosanoids; lipid signaling lysophosphatidic acid; metabolomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

