The age-phenome database
- PMID: 23984222
- PMCID: PMC3581109
- DOI: 10.1186/2193-1801-1-4
The age-phenome database
Abstract
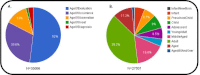
Data linking specific ages or age ranges with disease are abundant in biomedical literature. However, these data are organized such that searching for age-phenotype relationships is difficult. Recently, we described the Age-Phenome Knowledge-base (APK), a computational platform for storage and retrieval of information concerning age-related phenotypic patterns. Here, we report that data derived from over 1.5 million human-related PubMed abstracts have been added to APK. Using a text-mining pipeline, 35,683 entries which describe relationships between age and phenotype (such as disease) have been introduced into the database. Comparing the results to those obtained by a human reader reveals that the overall accuracy of these entries is estimated to exceed 80%. The usefulness of these data for obtaining new insight regarding age-disease relationships is demonstrated using clustering analysis, which is shown to capture obvious, as well as potentially interesting relationships between diseases. In addition, a new tool for browsing and searching the APK database is presented. We thus present a unique resource and a new framework for studying age-disease relationships and other phenotypic processes.
Keywords: Age; Knowledgebase; Phenotype; Text-minig.
Figures

References
-
- Diamond SG, Markham CH, Hoehn MM, McDowell FH, Muenter MD. Effect of age at onset on progression and mortality in Parkinson's disease. Neurology. 1989;39:1187. - PubMed
-
- Donaldson I, Martin J, de Bruijn B, Wolting C, Lay V, Tuekam B, Zhang S, Baskin B, Bader GD, Michalickova K, Pawson T, Hogue CW. PreBIND and Textomy-mining the biomedical literature for protein-protein interactions using a support vector machine. BMC Bioinformatics. 2003;4:11. doi: 10.1186/1471-2105-4-11. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

