Comparative analysis of context-dependent mutagenesis in humans and fruit flies
- PMID: 23984310
- PMCID: PMC3747623
- DOI: 10.1155/2013/173616
Comparative analysis of context-dependent mutagenesis in humans and fruit flies
Abstract
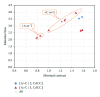
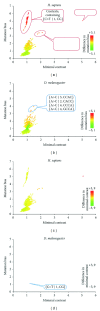
In general, mutation frequencies are context-dependent: specific adjacent nucleotides may influence the probability to observe a specific type of mutation in a genome. Recently, several hypermutable motifs were identified in the human genome. Namely, there is an increased frequency of T>C mutations in the second position of the words ATTG and ATAG and an increased frequency of A>C mutations in the first position of the word ACAA. Previous studies have also shown that there is a remarkable difference between the mutagenesis of humans and drosophila. While C>T mutations are overrepresented in the CG context in humans (and other vertebrates), this mutation regularity is not observed in Drosophila melanogaster. Such differences in the observed regularities of mutagenesis between representatives of different taxa might reflect differences in the mechanisms involved in mutagenesis. We performed a systematical comparison of mutation regularities within 2-4 bp contexts in Homo sapiens and Drosophila melanogaster and found that the aforementioned contexts are not hypermutable in fruit flies. It seems that most mutation contexts affect mutation rates in a similar manner in H. sapiens and D. melanogaster; however, several important exceptions are noted and discussed.
Figures

Similar articles
-
Comparative analysis of context-dependent mutagenesis using human and mouse models.Biomed Res Int. 2013;2013:989410. doi: 10.1155/2013/989410. Epub 2013 Aug 22. Biomed Res Int. 2013. PMID: 24058920 Free PMC article.
-
New words in human mutagenesis.BMC Bioinformatics. 2011 Jun 30;12:268. doi: 10.1186/1471-2105-12-268. BMC Bioinformatics. 2011. PMID: 21718472 Free PMC article.
-
Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster.Mol Biol Evol. 1988 Jul;5(4):393-414. doi: 10.1093/oxfordjournals.molbev.a040501. Mol Biol Evol. 1988. PMID: 3136295
-
Searching for sleep mutants of Drosophila melanogaster.Bioessays. 2003 Oct;25(10):940-9. doi: 10.1002/bies.10333. Bioessays. 2003. PMID: 14505361 Review.
-
Theoretical analysis of mutation hotspots and their DNA sequence context specificity.Mutat Res. 2003 Sep;544(1):65-85. doi: 10.1016/s1383-5742(03)00032-2. Mutat Res. 2003. PMID: 12888108 Review.
Cited by
-
Corrigendum to "Comparative Analysis of Context-Dependent Mutagenesis Using Human and Mouse Models".Biomed Res Int. 2020 Jul 20;2020:4657615. doi: 10.1155/2020/4657615. eCollection 2020. Biomed Res Int. 2020. PMID: 32775422 Free PMC article.
References
-
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nature Reviews Genetics. 2007;8(8):619–631. - PubMed
-
- Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrates genomes. Human Genetics. 1989;83(2):181–188. - PubMed
-
- Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408(6812):538–540. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

