Molecular footprints of domestication and improvement in soybean revealed by whole genome re-sequencing
- PMID: 23984715
- PMCID: PMC3844514
- DOI: 10.1186/1471-2164-14-579
Molecular footprints of domestication and improvement in soybean revealed by whole genome re-sequencing
Abstract
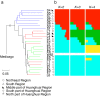
Background: Artificial selection played an important role in the origin of modern Glycine max cultivars from the wild soybean Glycine soja. To elucidate the consequences of artificial selection accompanying the domestication and modern improvement of soybean, 25 new and 30 published whole-genome re-sequencing accessions, which represent wild, domesticated landrace, and Chinese elite soybean populations were analyzed.
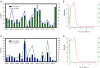
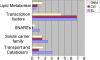
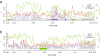
Results: A total of 5,102,244 single nucleotide polymorphisms (SNPs) and 707,969 insertion/deletions were identified. Among the SNPs detected, 25.5% were not described previously. We found that artificial selection during domestication led to more pronounced reduction in the genetic diversity of soybean than the switch from landraces to elite cultivars. Only a small proportion (2.99%) of the whole genomic regions appear to be affected by artificial selection for preferred agricultural traits. The selection regions were not distributed randomly or uniformly throughout the genome. Instead, clusters of selection hotspots in certain genomic regions were observed. Moreover, a set of candidate genes (4.38% of the total annotated genes) significantly affected by selection underlying soybean domestication and genetic improvement were identified.
Conclusions: Given the uniqueness of the soybean germplasm sequenced, this study drew a clear picture of human-mediated evolution of the soybean genomes. The genomic resources and information provided by this study would also facilitate the discovery of genes/loci underlying agronomically important traits.
Figures


References
-
- Hymowitz T. In: Soybeans: Improvement, Production and Uses. 3. Boerma HR, Specht JE, editor. Wisconsin, USA: Madison; 2004. Speciation and cytogenetics; pp. 97–129.
-
- Singh RJ, Hymowitz T. Soybean genetic resources and crop improvement. Genome. 1999;42:605–616. doi: 10.1139/g99-039. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

