Genome-wide gene expression profiling of stress response in a spinal cord clip compression injury model
- PMID: 23984903
- PMCID: PMC3846681
- DOI: 10.1186/1471-2164-14-583
Genome-wide gene expression profiling of stress response in a spinal cord clip compression injury model
Abstract
Background: The aneurysm clip impact-compression model of spinal cord injury (SCI) is a standard injury model in animals that closely mimics the primary mechanism of most human injuries: acute impact and persisting compression. Its histo-pathological and behavioural outcomes are extensively similar to human SCI. To understand the distinct molecular events underlying this injury model we analyzed global mRNA abundance changes during the acute, subacute and chronic stages of a moderate to severe injury to the rat spinal cord.
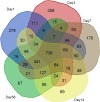
Results: Time-series expression analyses resulted in clustering of the majority of deregulated transcripts into eight statistically significant expression profiles. Systematic application of Gene Ontology (GO) enrichment pathway analysis allowed inference of biological processes participating in SCI pathology. Temporal analysis identified events specific to and common between acute, subacute and chronic time-points. Processes common to all phases of injury include blood coagulation, cellular extravasation, leukocyte cell-cell adhesion, the integrin-mediated signaling pathway, cytokine production and secretion, neutrophil chemotaxis, phagocytosis, response to hypoxia and reactive oxygen species, angiogenesis, apoptosis, inflammatory processes and ossification. Importantly, various elements of adaptive and induced innate immune responses span, not only the acute and subacute phases, but also persist throughout the chronic phase of SCI. Induced innate responses, such as Toll-like receptor signaling, are more active during the acute phase but persist throughout the chronic phase. However, adaptive immune response processes such as B and T cell activation, proliferation, and migration, T cell differentiation, B and T cell receptor-mediated signaling, and B cell- and immunoglobulin-mediated immune response become more significant during the chronic phase.
Conclusions: This analysis showed that, surprisingly, the diverse series of molecular events that occur in the acute and subacute stages persist into the chronic stage of SCI. The strong agreement between our results and previous findings suggest that our analytical approach will be useful in revealing other biological processes and genes contributing to SCI pathology.
Figures





Similar articles
-
Transcriptome profile of rat genes in injured spinal cord at different stages by RNA-sequencing.BMC Genomics. 2017 Feb 15;18(1):173. doi: 10.1186/s12864-017-3532-x. BMC Genomics. 2017. PMID: 28201982 Free PMC article.
-
Investigation of candidate long noncoding RNAs and messenger RNAs in the immediate phase of spinal cord injury based on gene expression profiles.Gene. 2018 Jun 30;661:119-125. doi: 10.1016/j.gene.2018.03.074. Epub 2018 Mar 23. Gene. 2018. PMID: 29580899
-
Integrated Bioinformatics Analysis of Hub Genes and Pathways Associated with a Compression Model of Spinal Cord Injury in Rats.Med Sci Monit. 2020 Nov 5;26:e927107. doi: 10.12659/MSM.927107. Med Sci Monit. 2020. PMID: 33149108 Free PMC article.
-
Experimental spinal cord trauma: a review of mechanically induced spinal cord injury in rat models.Rev Neurosci. 2017 Jan 1;28(1):15-20. doi: 10.1515/revneuro-2016-0050. Rev Neurosci. 2017. PMID: 27845888 Review.
-
Oxidative DNA Damage in the Pathophysiology of Spinal Cord Injury: Seems Obvious, but Where Is the Evidence?Antioxidants (Basel). 2022 Aug 31;11(9):1728. doi: 10.3390/antiox11091728. Antioxidants (Basel). 2022. PMID: 36139802 Free PMC article. Review.
Cited by
-
Activating Transcription Factor 3 (ATF3) is a Highly Conserved Pro-regenerative Transcription Factor in the Vertebrate Nervous System.Front Cell Dev Biol. 2022 Mar 8;10:824036. doi: 10.3389/fcell.2022.824036. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35350379 Free PMC article. Review.
-
Chronic spinal cord changes in a high-fat diet-fed male rat model of thoracic spinal contusion.Physiol Genomics. 2017 Sep 1;49(9):519-529. doi: 10.1152/physiolgenomics.00078.2017. Epub 2017 Aug 18. Physiol Genomics. 2017. PMID: 28821567 Free PMC article.
-
Brainstem-Evoked Transcription of Defensive Genes After Spinal Cord Injury.Front Cell Neurosci. 2019 Nov 19;13:510. doi: 10.3389/fncel.2019.00510. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31803022 Free PMC article.
-
T cell deficiency in spinal cord injury: altered locomotor recovery and whole-genome transcriptional analysis.BMC Neurosci. 2015 Nov 6;16:74. doi: 10.1186/s12868-015-0212-0. BMC Neurosci. 2015. PMID: 26546062 Free PMC article.
-
Bibliometric analysis of research on gene expression in spinal cord injury.Front Mol Neurosci. 2022 Oct 31;15:1023692. doi: 10.3389/fnmol.2022.1023692. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36385766 Free PMC article.
References
-
- Watson BD, Prado R, Dietrich WD, Ginsberg MD, Green BA. Photochemically induced spinal cord injury in the rat. Brain Res. 1986;367(1–2):296–300. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

