Inferring the expression variability of human transposable element-derived exons by linear model analysis of deep RNA sequencing data
- PMID: 23984937
- PMCID: PMC3765721
- DOI: 10.1186/1471-2164-14-584
Inferring the expression variability of human transposable element-derived exons by linear model analysis of deep RNA sequencing data
Abstract
Background: The exonization of transposable elements (TEs) has proven to be a significant mechanism for the creation of novel exons. Existing knowledge of the retention patterns of TE exons in mRNAs were mainly established by the analysis of Expressed Sequence Tag (EST) data and microarray data.
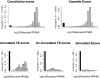
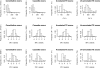
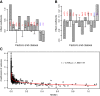
Results: This study seeks to validate and extend previous studies on the expression of TE exons by an integrative statistical analysis of high throughput RNA sequencing data. We collected 26 RNA-seq datasets spanning multiple tissues and cancer types. The exon-level digital expressions (indicating retention rates in mRNAs) were quantified by a double normalized measure, called the rescaled RPKM (Reads Per Kilobase of exon model per Million mapped reads). We analyzed the distribution profiles and the variability (across samples and between tissue/disease groups) of TE exon expressions, and compared them with those of other constitutive or cassette exons. We inferred the effects of four genomic factors, including the location, length, cognate TE family and TE nucleotide proportion (RTE, see Methods section) of a TE exon, on the exons' expression level and expression variability. We also investigated the biological implications of an assembly of highly-expressed TE exons.
Conclusion: Our analysis confirmed prior studies from the following four aspects. First, with relatively high expression variability, most TE exons in mRNAs, especially those without exact counterparts in the UCSC RefSeq (Reference Sequence) gene tables, demonstrate low but still detectable expression levels in most tissue samples. Second, the TE exons in coding DNA sequences (CDSs) are less highly expressed than those in 3' (5') untranslated regions (UTRs). Third, the exons derived from chronologically ancient repeat elements, such as MIRs, tend to be highly expressed in comparison with those derived from younger TEs. Fourth, the previously observed negative relationship between the lengths of exons and the inclusion levels in transcripts is also true for exonized TEs. Furthermore, our study resulted in several novel findings. They include: (1) for the TE exons with non-zero expression and as shown in most of the studied biological samples, a high TE nucleotide proportion leads to their lower retention rates in mRNAs; (2) the considered genomic features (i.e. a continuous variable such as the exon length or a category indicator such as 3'UTR) influence the expression level and the expression variability (CV) of TE exons in an inverse manner; (3) not only the exons derived from Alu elements but also the exons from the TEs of other families were preferentially established in zinc finger (ZNF) genes.
Figures


Similar articles
-
ExoPLOT: Representation of alternative splicing in human tissues and developmental stages with transposed element (TE) involvement.Genomics. 2022 Jul;114(4):110434. doi: 10.1016/j.ygeno.2022.110434. Epub 2022 Jul 18. Genomics. 2022. PMID: 35863675
-
Evaluating the protein coding potential of exonized transposable element sequences.Biol Direct. 2007 Nov 26;2:31. doi: 10.1186/1745-6150-2-31. Biol Direct. 2007. PMID: 18036258 Free PMC article.
-
Transposable elements in disease-associated cryptic exons.Hum Genet. 2010 Feb;127(2):135-54. doi: 10.1007/s00439-009-0752-4. Epub 2009 Oct 10. Hum Genet. 2010. PMID: 19823873
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
-
The intertwining of transposable elements and non-coding RNAs.Int J Mol Sci. 2013 Jun 26;14(7):13307-28. doi: 10.3390/ijms140713307. Int J Mol Sci. 2013. PMID: 23803660 Free PMC article. Review.
Cited by
-
Transcriptome profiling of the cancer and adjacent nontumor tissues from cervical squamous cell carcinoma patients by RNA sequencing.Tumour Biol. 2015 May;36(5):3309-17. doi: 10.1007/s13277-014-2963-0. Epub 2015 Jan 14. Tumour Biol. 2015. PMID: 25586346
-
Regulation of human interferon signaling by transposon exonization.Cell. 2024 Dec 26;187(26):7621-7636.e19. doi: 10.1016/j.cell.2024.11.016. Epub 2024 Dec 12. Cell. 2024. PMID: 39672162
-
De novo sequencing, assembly and analysis of eight different transcriptomes from the Malayan pangolin.Sci Rep. 2016 Sep 13;6:28199. doi: 10.1038/srep28199. Sci Rep. 2016. PMID: 27618997 Free PMC article.
-
Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates.Chromosome Res. 2015 Sep;23(3):505-31. doi: 10.1007/s10577-015-9493-5. Chromosome Res. 2015. PMID: 26395902 Review.
References
-
- Affymetrix website. http://www.affymetrix.com/estore/browse/products.jsp?productId=131452#1_1.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

