Identification of Xin-repeat proteins as novel ligands of the SH3 domains of nebulin and nebulette and analysis of their interaction during myofibril formation and remodeling
- PMID: 23985323
- PMCID: PMC3810769
- DOI: 10.1091/mbc.E13-04-0202
Identification of Xin-repeat proteins as novel ligands of the SH3 domains of nebulin and nebulette and analysis of their interaction during myofibril formation and remodeling
Abstract
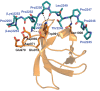
The Xin actin-binding repeat-containing proteins Xin and XIRP2 are exclusively expressed in striated muscle cells, where they are believed to play an important role in development. In adult muscle, both proteins are concentrated at attachment sites of myofibrils to the membrane. In contrast, during development they are localized to immature myofibrils together with their binding partner, filamin C, indicating an involvement of both proteins in myofibril assembly. We identify the SH3 domains of nebulin and nebulette as novel ligands of proline-rich regions of Xin and XIRP2. Precise binding motifs are mapped and shown to bind both SH3 domains with micromolar affinity. Cocrystallization of the nebulette SH3 domain with the interacting XIRP2 peptide PPPTLPKPKLPKH reveals selective interactions that conform to class II SH3 domain-binding peptides. Bimolecular fluorescence complementation experiments in cultured muscle cells indicate a temporally restricted interaction of Xin-repeat proteins with nebulin/nebulette during early stages of myofibril development that is lost upon further maturation. In mature myofibrils, this interaction is limited to longitudinally oriented structures associated with myofibril development and remodeling. These data provide new insights into the role of Xin actin-binding repeat-containing proteins (together with their interaction partners) in myofibril assembly and after muscle damage.
Figures






Similar articles
-
Functional dissection of nebulette demonstrates actin binding of nebulin-like repeats and Z-line targeting of SH3 and linker domains.Cell Motil Cytoskeleton. 1999;44(1):1-22. doi: 10.1002/(SICI)1097-0169(199909)44:1<1::AID-CM1>3.0.CO;2-8. Cell Motil Cytoskeleton. 1999. PMID: 10470015
-
Aciculin interacts with filamin C and Xin and is essential for myofibril assembly, remodeling and maintenance.J Cell Sci. 2014 Aug 15;127(Pt 16):3578-92. doi: 10.1242/jcs.152157. Epub 2014 Jun 24. J Cell Sci. 2014. PMID: 24963132
-
Zyxin interacts with the SH3 domains of the cytoskeletal proteins LIM-nebulette and Lasp-1.J Biol Chem. 2004 May 7;279(19):20401-10. doi: 10.1074/jbc.M310304200. Epub 2004 Mar 5. J Biol Chem. 2004. PMID: 15004028
-
Roles of Nebulin Family Members in the Heart.Circ J. 2015;79(10):2081-7. doi: 10.1253/circj.CJ-15-0854. Epub 2015 Aug 31. Circ J. 2015. PMID: 26321576 Review.
-
New insights into the roles of Xin repeat-containing proteins in cardiac development, function, and disease.Int Rev Cell Mol Biol. 2014;310:89-128. doi: 10.1016/B978-0-12-800180-6.00003-7. Int Rev Cell Mol Biol. 2014. PMID: 24725425 Free PMC article. Review.
Cited by
-
Nebulin: big protein with big responsibilities.J Muscle Res Cell Motil. 2020 Mar;41(1):103-124. doi: 10.1007/s10974-019-09565-3. Epub 2020 Jan 25. J Muscle Res Cell Motil. 2020. PMID: 31982973 Free PMC article. Review.
-
A survey of transcriptome complexity in Sus scrofa using single-molecule long-read sequencing.DNA Res. 2018 Aug 1;25(4):421-437. doi: 10.1093/dnares/dsy014. DNA Res. 2018. PMID: 29850846 Free PMC article.
-
Computed Protein-Protein Enthalpy Signatures as a Tool for Identifying Conformation Sampling Problems.J Chem Inf Model. 2023 Oct 9;63(19):6095-6108. doi: 10.1021/acs.jcim.3c01041. Epub 2023 Sep 27. J Chem Inf Model. 2023. PMID: 37759363 Free PMC article.
-
Deleting nebulin's C-terminus reveals its importance to sarcomeric structure and function and is sufficient to invoke nemaline myopathy.Hum Mol Genet. 2019 May 15;28(10):1709-1725. doi: 10.1093/hmg/ddz016. Hum Mol Genet. 2019. PMID: 30689900 Free PMC article.
-
Comprehensive Transcriptome Analysis of Sex-Biased Expressed Genes Reveals Discrete Biological and Physiological Features of Male and Female Schistosoma japonicum.PLoS Negl Trop Dis. 2016 Apr 29;10(4):e0004684. doi: 10.1371/journal.pntd.0004684. eCollection 2016 Apr. PLoS Negl Trop Dis. 2016. PMID: 27128440 Free PMC article.
References
-
- Carlsson L, Yu JG, Thornell LE. New aspects of obscurin in human striated muscles. Histochem Cell Biol. 2008;130:91–103. - PubMed
-
- Claeys KG, et al. Differential involvement of sarcomeric proteins in myofibrillar myopathies: a morphological and immunohistochemical study. Acta Neuropathol. 2009;117:293–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

