Phylogenomic data support a seventh order of Methylotrophic methanogens and provide insights into the evolution of Methanogenesis
- PMID: 23985970
- PMCID: PMC3814188
- DOI: 10.1093/gbe/evt128
Phylogenomic data support a seventh order of Methylotrophic methanogens and provide insights into the evolution of Methanogenesis
Abstract
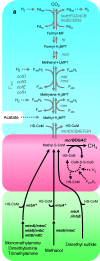
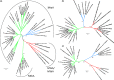
Increasing evidence from sequence data from various environments, including the human gut, suggests the existence of a previously unknown putative seventh order of methanogens. The first genomic data from members of this lineage, Methanomassiliicoccus luminyensis and "Candidatus Methanomethylophilus alvus," provide insights into its evolutionary history and metabolic features. Phylogenetic analysis of ribosomal proteins robustly indicates a monophyletic group independent of any previously known methanogenic order, which shares ancestry with the Marine Benthic Group D, the Marine Group II, the DHVE2 group, and the Thermoplasmatales. This phylogenetic position, along with the analysis of enzymes involved in core methanogenesis, strengthens a single ancient origin of methanogenesis in the Euryarchaeota and indicates further multiple independent losses of this metabolism in nonmethanogenic lineages than previously suggested. Genomic analysis revealed an unprecedented loss of the genes coding for the first six steps of methanogenesis from H₂/CO₂ and the oxidative part of methylotrophic methanogenesis, consistent with the fact that M. luminyensis and "Ca. M. alvus" are obligate H₂-dependent methylotrophic methanogens. Genomic data also suggest that these methanogens may use a large panel of methylated compounds. Phylogenetic analysis including homologs retrieved from environmental samples indicates that methylotrophic methanogenesis (regardless of dependency on H₂) is not restricted to gut representatives but may be an ancestral characteristic of the whole order, and possibly also of ancient origin in the Euryarchaeota. 16S rRNA and McrA trees show that this new order of methanogens is very diverse and occupies environments highly relevant for methane production, therefore representing a key lineage to fully understand the diversity and evolution of methanogenesis.
Keywords: Archaea; Methanomassiliicoccales; Methanoplasmatales; evolution; genomics; methanogenesis.
Figures



References
-
- Biderre-Petit C, et al. Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol Ecol. 2011;77:533–545. - PubMed
-
- Brochier-Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol. 2011;14:274–281. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

