Engineered protease-resistant antibodies with selectable cell-killing functions
- PMID: 23986451
- PMCID: PMC3829399
- DOI: 10.1074/jbc.M113.486142
Engineered protease-resistant antibodies with selectable cell-killing functions
Abstract
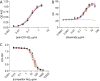
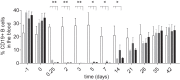
Molecularly engineered antibodies with fit-for-purpose properties will differentiate next generation antibody therapeutics from traditional IgG1 scaffolds. One requirement for engineering the most appropriate properties for a particular therapeutic area is an understanding of the intricacies of the target microenvironment in which the antibody is expected to function. Our group and others have demonstrated that proteases secreted by invasive tumors and pathological microorganisms are capable of cleaving human IgG1, the most commonly adopted isotype among monoclonal antibody therapeutics. Specific cleavage in the lower hinge of IgG1 results in a loss of Fc-mediated cell-killing functions without a concomitant loss of antigen binding capability or circulating antibody half-life. Proteolytic cleavage in the hinge region by tumor-associated or microbial proteases is postulated as a means of evading host immune responses, and antibodies engineered with potent cell-killing functions that are also resistant to hinge proteolysis are of interest. Mutation of the lower hinge region of an IgG1 resulted in protease resistance but also resulted in a profound loss of Fc-mediated cell-killing functions. In the present study, we demonstrate that specific mutations of the CH2 domain in conjunction with lower hinge mutations can restore and sometimes enhance cell-killing functions while still retaining protease resistance. By identifying mutations that can restore either complement- or Fcγ receptor-mediated functions on a protease-resistant scaffold, we were able to generate a novel protease-resistant platform with selective cell-killing functionality.
Keywords: Antibody Engineering; Immunology; Innate Immunity; Macrophages; NK Cells.
Figures


References
-
- Presta L. G. (2008) Molecular engineering and design of therapeutic antibodies. Curr. Opin. Immunol. 20, 460–470 - PubMed
-
- Strohl W. R. (2009) Optimization of Fc-mediated effector functions of monoclonal antibodies. Curr. Opin. Biotechnol. 20, 685–691 - PubMed
-
- Idusogie E. E., Wong P. Y., Presta L. G., Gazzano-Santoro H., Totpal K., Ultsch M., Mulkerrin M. G. (2001) Engineered antibodies with increased activity to recruit complement. J. Immunol. 166, 2571–2575 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

