Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex
- PMID: 23986495
- PMCID: PMC3780858
- DOI: 10.1073/pnas.1305687110
Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex
Abstract
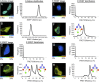
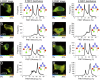
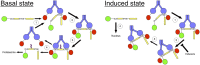
The transcription factor NF-E2 p45-related factor 2 (Nrf2), a master regulator of cytoprotective genes, is controlled by dimeric Kelch-like ECH associated protein 1 (Keap1), a substrate adaptor protein for Cullin3/RING-box protein 1 ubiquitin ligase, which normally targets Nrf2 for ubiquitination and degradation but loses this ability in response to electrophiles and oxidants (inducers). By using recombinant proteins and populations of cells, some of the general features of the regulation of Nrf2 by Keap1 have been outlined. However, how the two proteins interact at a single-cell level is presently unknown. We now report the development of a quantitative Förster resonance energy transfer-based system using multiphoton fluorescence lifetime imaging microscopy and its application for investigating the interaction between Nrf2 and Keap1 in single live cells. By using this approach, we found that under homeostatic conditions, the interaction between Keap1 and Nrf2 follows a cycle in which the complex sequentially adopts two distinct conformations: "open," in which Nrf2 interacts with a single molecule of Keap1, followed by "closed," in which Nrf2 binds to both members of the Keap1 dimer. Inducers disrupt this cycle by causing accumulation of the complex in the closed conformation without release of Nrf2. As a consequence, free Keap1 is not regenerated, and newly synthesized Nrf2 is stabilized. On the basis of these findings, we propose a model we have named the "cyclic sequential attachment and regeneration model of Keap1-mediated degradation of Nrf2." This previously unanticipated dynamism allows rapid transcriptional responses to environmental changes and can accommodate multiple modes of regulation.
Keywords: FLIM; FRET; protein–protein interactions; sulforaphane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

