The open chromatin landscape of Kaposi's sarcoma-associated herpesvirus
- PMID: 23986576
- PMCID: PMC3807352
- DOI: 10.1128/JVI.01685-13
The open chromatin landscape of Kaposi's sarcoma-associated herpesvirus
Abstract
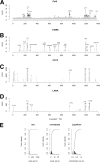
Kaposi's sarcoma-associated herpesvirus (KSHV) is an oncogenic gammaherpesvirus which establishes latent infection in endothelial and B cells, as well as in primary effusion lymphoma (PEL). During latency, the viral genome exists as a circular DNA minichromosome (episome) and is packaged into chromatin analogous to human chromosomes. Only a small subset of promoters, those which drive latent RNAs, are active in latent episomes. In general, nucleosome depletion ("open chromatin") is a hallmark of eukaryotic regulatory elements such as promoters and transcriptional enhancers or insulators. We applied formaldehyde-assisted isolation of regulatory elements (FAIRE) followed by next-generation sequencing to identify regulatory elements in the KSHV genome and integrated these data with previously identified locations of histone modifications, RNA polymerase II occupancy, and CTCF binding sites. We found that (i) regions of open chromatin were not restricted to the transcriptionally defined latent loci; (ii) open chromatin was adjacent to regions harboring activating histone modifications, even at transcriptionally inactive loci; and (iii) CTCF binding sites fell within regions of open chromatin with few exceptions, including the constitutive LANA promoter and the vIL6 promoter. FAIRE-identified nucleosome depletion was similar among B and endothelial cell lineages, suggesting a common viral genome architecture in all forms of latency.
Figures





References
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 - PubMed
-
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186–1191 - PubMed
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA019014/CA/NCI NIH HHS/United States
- R01 CA109232/CA/NCI NIH HHS/United States
- T32 AI007419/AI/NIAID NIH HHS/United States
- CA109232/CA/NCI NIH HHS/United States
- T32 GM007092/GM/NIGMS NIH HHS/United States
- CA019014/CA/NCI NIH HHS/United States
- T32GM067553/GM/NIGMS NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- T32 GM067553/GM/NIGMS NIH HHS/United States
- CA166447/CA/NCI NIH HHS/United States
- AI107810/AI/NIAID NIH HHS/United States
- R01 CA166447/CA/NCI NIH HHS/United States
- R01 DE018304/DE/NIDCR NIH HHS/United States
- U19 AI107810/AI/NIAID NIH HHS/United States
- T32AI007419/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

