A cholesterol recognition amino acid consensus domain in GP64 fusion protein facilitates anchoring of baculovirus to mammalian cells
- PMID: 23986592
- PMCID: PMC3807332
- DOI: 10.1128/JVI.01356-13
A cholesterol recognition amino acid consensus domain in GP64 fusion protein facilitates anchoring of baculovirus to mammalian cells
Abstract
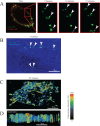
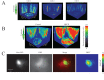
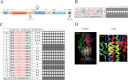
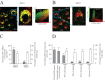
Baculoviridae is a large family of double-stranded DNA viruses that selectively infect insects. Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the best-studied baculovirus from the family. Many studies over the last several years have shown that AcMNPV can enter a wide variety of mammalian cells and deliver genetic material for foreign gene expression. While most animal viruses studied so far have developed sophisticated mechanisms to selectively infect specific cells and tissues in an organism, AcMNPV can penetrate and deliver foreign genes into most cells studied to this date. The details about the mechanisms of internalization have been partially described. In the present study, we have identified a cholesterol recognition amino acid consensus (CRAC) domain present in the AcMNPV envelope fusion protein GP64. We demonstrated the association of a CRAC domain with cholesterol, which is important to facilitate the anchoring of the virus at the mammalian cell membrane. Furthermore, this initial anchoring favors AcMNPV endocytosis via a dynamin- and clathrin-dependent mechanism. Under these conditions, efficient baculovirus-driven gene expression is obtained. In contrast, when cholesterol is reduced from the plasma membrane, AcMNPV enters the cell via a dynamin- and clathrin-independent mechanism. The result of using this alternative internalization pathway is a reduced level of baculovirus-driven gene expression. This study is the first to document the importance of a novel CRAC domain in GP64 and its role in modulating gene delivery in AcMNPV.
Figures



References
-
- Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Thiem SM, Vlak JM. 2006. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 151:1257–1266 - PubMed
-
- Caron AW, Archambault J, Massie B. 1990. High-level recombinant protein production in bioreactors using the baculovirus-insect cell expression system. Biotechnol. Bioeng. 36:1133–1140 - PubMed
-
- Elias CB, Jardin B, Kamen A. 2007. Recombinant protein production in large-scale agitated bioreactors using the baculovirus expression vector system. Methods Mol. Biol. 388:225–246 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

