Neutron diffraction studies towards deciphering the protonation state of catalytic residues in the bacterial KDN9P phosphatase
- PMID: 23989152
- PMCID: PMC3758152
- DOI: 10.1107/S1744309113021386
Neutron diffraction studies towards deciphering the protonation state of catalytic residues in the bacterial KDN9P phosphatase
Abstract
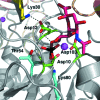
The enzyme 2-keto-3-deoxy-9-O-phosphonononic acid phosphatase (KDN9P phosphatase) functions in the pathway for the production of 2-keto-3-deoxy-D-glycero-D-galacto-nononic acid, a sialic acid that is important for the survival of commensal bacteria in the human intestine. The enzyme is a member of the haloalkanoate dehalogenase superfamily and represents a good model for the active-site protonation state of family members. Crystals of approximate dimensions 1.5 × 1.0 × 1.0 mm were obtained in space group P2(1)2(1)2, with unit-cell parameters a = 83.1, b = 108.9, c = 75.7 Å. A complete neutron data set was collected from a medium-sized H/D-exchanged crystal at BIODIFF at the Heinz Maier-Leibnitz Zentrum (MLZ), Garching, Germany in 18 d. Initial refinement to 2.3 Å resolution using only neutron data showed significant density for catalytically important residues.
Keywords: KDN9P phosphatase; neutron diffraction; protonation states.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

