The MaSuRCA genome assembler
- PMID: 23990416
- PMCID: PMC3799473
- DOI: 10.1093/bioinformatics/btt476
The MaSuRCA genome assembler
Abstract
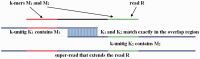
Motivation: Second-generation sequencing technologies produce high coverage of the genome by short reads at a low cost, which has prompted development of new assembly methods. In particular, multiple algorithms based on de Bruijn graphs have been shown to be effective for the assembly problem. In this article, we describe a new hybrid approach that has the computational efficiency of de Bruijn graph methods and the flexibility of overlap-based assembly strategies, and which allows variable read lengths while tolerating a significant level of sequencing error. Our method transforms large numbers of paired-end reads into a much smaller number of longer 'super-reads'. The use of super-reads allows us to assemble combinations of Illumina reads of differing lengths together with longer reads from 454 and Sanger sequencing technologies, making it one of the few assemblers capable of handling such mixtures. We call our system the Maryland Super-Read Celera Assembler (abbreviated MaSuRCA and pronounced 'mazurka').
Results: We evaluate the performance of MaSuRCA against two of the most widely used assemblers for Illumina data, Allpaths-LG and SOAPdenovo2, on two datasets from organisms for which high-quality assemblies are available: the bacterium Rhodobacter sphaeroides and chromosome 16 of the mouse genome. We show that MaSuRCA performs on par or better than Allpaths-LG and significantly better than SOAPdenovo on these data, when evaluated against the finished sequence. We then show that MaSuRCA can significantly improve its assemblies when the original data are augmented with long reads.
Availability: MaSuRCA is available as open-source code at ftp://ftp.genome.umd.edu/pub/MaSuRCA/. Previous (pre-publication) releases have been publicly available for over a year.
Contact: alekseyz@ipst.umd.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

