Integrating milk metabolite profile information for the prediction of traditional milk traits based on SNP information for Holstein cows
- PMID: 23990900
- PMCID: PMC3749218
- DOI: 10.1371/journal.pone.0070256
Integrating milk metabolite profile information for the prediction of traditional milk traits based on SNP information for Holstein cows
Abstract
In this study the benefit of metabolome level analysis for the prediction of genetic value of three traditional milk traits was investigated. Our proposed approach consists of three steps: First, milk metabolite profiles are used to predict three traditional milk traits of 1,305 Holstein cows. Two regression methods, both enabling variable selection, are applied to identify important milk metabolites in this step. Second, the prediction of these important milk metabolite from single nucleotide polymorphisms (SNPs) enables the detection of SNPs with significant genetic effects. Finally, these SNPs are used to predict milk traits. The observed precision of predicted genetic values was compared to the results observed for the classical genotype-phenotype prediction using all SNPs or a reduced SNP subset (reduced classical approach). To enable a comparison between SNP subsets, a special invariable evaluation design was implemented. SNPs close to or within known quantitative trait loci (QTL) were determined. This enabled us to determine if detected important SNP subsets were enriched in these regions. The results show that our approach can lead to genetic value prediction, but requires less than 1% of the total amount of (40,317) SNPs., significantly more important SNPs in known QTL regions were detected using our approach compared to the reduced classical approach. Concluding, our approach allows a deeper insight into the associations between the different levels of the genotype-phenotype map (genotype-metabolome, metabolome-phenotype, genotype-phenotype).
Conflict of interest statement
Figures

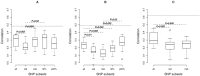
 = 0.05), this is marked with a black dashed line and the observed P-value is given. The gray line represents a possible upper bound for the accuracy of prediction given as the square root of the estimated narrow-sense heritability based on a sire model.
= 0.05), this is marked with a black dashed line and the observed P-value is given. The gray line represents a possible upper bound for the accuracy of prediction given as the square root of the estimated narrow-sense heritability based on a sire model.References
-
- Geishauser T, Leslie K, Tenhag J, Bashiri A (2000) Evaluation of eight cow-side ketone tests in milk for detection of subclinical ketosis in dairy cows. J Dairy Sci 83: 296–299. - PubMed
-
- Klein MS, Buttchereit N, Miemczyk SP, Immervoll AK, Louis C, et al. (2012) NMR metabolomic analysis of dairy cows reveals milk glycerophosphocholine to phosphocholine ratio as prognostic biomarker for risk of ketosis. J Proteome Res 11: 1373–1381. - PubMed
-
- Cabrita ARJ, Fonseca AJM, Dewhurst RJ, Gomes E (2003) Nitrogen supplementation of corn silages. 2. Assessing rumen function using fatty acid profiles of bovine milk. J Dairy Sci 86: 4020–4032. - PubMed
-
- Farr V, Prosser C, Nicholas G, Lee J, Hart A, et al. (2002) Increased milk lactic acid concentration is an early indicator of mastitis. New Zealand Society of Animal Production, volume 62 22–23.
-
- Hastie T, Tibshirani R, Friedman J (2009) The elements of statistical learning: data mining, inference and prediction. Springer, 2 edition.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

