Non contiguous-finished genome sequence and description of Dielma fastidiosa gen. nov., sp. nov., a new member of the Family Erysipelotrichaceae
- PMID: 23991263
- PMCID: PMC3746426
- DOI: 10.4056/sigs.3567059
Non contiguous-finished genome sequence and description of Dielma fastidiosa gen. nov., sp. nov., a new member of the Family Erysipelotrichaceae
Abstract
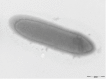
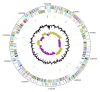
Dielma fastidiosa strain JC13(T) gen. nov., sp. nov. is the type strain of D. fastidiosa gen. nov., sp. nov., the type species of a new genus within the family Erysipelotrichaceae. This strain, whose draft genome is described here, was isolated from the fecal flora of a healthy 16-year-old male Senegalese volunteer. D. fastidiosa is a Gram-negative anaerobic rod. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 3,574,031 bp long genome comprises a 3,556,241-bp chromosome and a 17,790-bp plasmid. The chromosome contains 3,441 protein-coding and 50 RNA genes, including 3 rRNA genes, whereas the plasmid contains 17 protein-coding genes.
Keywords: Culturomics; Dielma fastidiosa; Genome; Taxono-genomics.
Figures


References
-
- Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 2012; 18:1185-1193 - PubMed
-
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 2006; 33:152-155
-
- Wayne LG, Brenner DJ, Colwell RR, Grimont AD, Kandler O, Krichevsky MI, Moore LH, Moore EC, Murray GE, Sktackbrandt E, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 1987; 37:463-464 10.1099/00207713-37-4-463 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

