Molecular insight into substrate recognition and catalysis of Baeyer-Villiger monooxygenase MtmOIV, the key frame-modifying enzyme in the biosynthesis of anticancer agent mithramycin
- PMID: 23992662
- PMCID: PMC3830731
- DOI: 10.1021/cb400399b
Molecular insight into substrate recognition and catalysis of Baeyer-Villiger monooxygenase MtmOIV, the key frame-modifying enzyme in the biosynthesis of anticancer agent mithramycin
Abstract
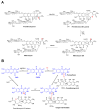
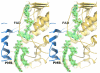
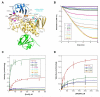
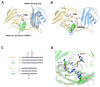
Baeyer-Villiger monooxygenases (BVMOs) have been shown to play key roles for the biosynthesis of important natural products. MtmOIV, a homodimeric FAD- and NADPH-dependent BVMO, catalyzes the key frame-modifying steps of the mithramycin biosynthetic pathway, including an oxidative C-C bond cleavage, by converting its natural substrate premithramycin B into mithramycin DK, the immediate precursor of mithramycin. The drastically improved protein structure of MtmOIV along with the high-resolution structure of MtmOIV in complex with its natural substrate premithramycin B are reported here, revealing previously undetected key residues that are important for substrate recognition and catalysis. Kinetic analyses of selected mutants allowed us to probe the substrate binding pocket of MtmOIV and also to discover the putative NADPH binding site. This is the first substrate-bound structure of MtmOIV providing new insights into substrate recognition and catalysis, which paves the way for the future design of a tailored enzyme for the chemo-enzymatic preparation of novel mithramycin analogues.
Figures



References
-
- Leisch H, Morley K, Lau PC. Baeyer-Villiger monooxygenases: more than just green chemistry. Chem. Rev. 2011;111:4165–4222. - PubMed
-
- Dover LG, Corsino PE, Daniels IR, Cocklin SL, Tatituri V, Besra GS, Futterer K. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 2004;340:1095–1105. - PubMed
-
- Koma D, Sakashita Y, Kubota K, Fujii Y, Hasumi F, Chung SY, Kubo M. Degradation pathways of cyclic alkanes in Rhodococcus sp. NDKK48. Appl. Microbiol. Biotechnol. 2004;66:92–99. - PubMed
-
- Leisch H, Shi R, Grosse S, Morley K, Bergeron H, Cygler M, Iwaki H, Hasegawa Y, Lau PC. Camphor Pathway 2-Oxo-{Delta}3-4,5,5-trimethylcyclopentenylacetyl-CoA Monooxygenase of Pseudomonas putida ATCC 17453: Cloning, Baeyer-Villiger Biooxidations, and Structures. Appl. Environ. Microbiol. 2012;78:2200–2212. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

