Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma
- PMID: 23993863
- PMCID: PMC3817560
- DOI: 10.1016/j.ccr.2013.08.001
Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma
Abstract
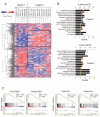
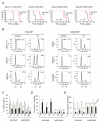
Despite extensive study, few therapeutic targets have been identified for glioblastoma (GBM). Here we show that patient-derived glioma sphere cultures (GSCs) that resemble either the proneural (PN) or mesenchymal (MES) transcriptomal subtypes differ significantly in their biological characteristics. Moreover, we found that a subset of the PN GSCs undergoes differentiation to a MES state in a TNF-α/NF-κB-dependent manner with an associated enrichment of CD44 subpopulations and radioresistant phenotypes. We present data to suggest that the tumor microenvironment cell types such as macrophages/microglia may play an integral role in this process. We further show that the MES signature, CD44 expression, and NF-κB activation correlate with poor radiation response and shorter survival in patients with GBM.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures






References
-
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. - PubMed
-
- Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. - PubMed
-
- Bredel M, Bredel C, Juric D, Duran GE, Yu RX, Harsh GR, Vogel H, Recht LD, Scheck AC, Sikic BI. Tumor necrosis factor-alpha-induced protein 3 as a putative regulator of nuclear factor-kappaB-mediated resistance to O6-alkylating agents in human glioblastomas. J Clin Oncol. 2006;24:274–287. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
