The regulation of mitochondrial DNA copy number in glioblastoma cells
- PMID: 23995230
- PMCID: PMC3824586
- DOI: 10.1038/cdd.2013.115
The regulation of mitochondrial DNA copy number in glioblastoma cells
Abstract
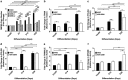
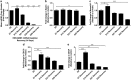
As stem cells undergo differentiation, mitochondrial DNA (mtDNA) copy number is strictly regulated in order that specialized cells can generate appropriate levels of adenosine triphosphate (ATP) through oxidative phosphorylation (OXPHOS) to undertake their specific functions. It is not understood whether tumor-initiating cells regulate their mtDNA in a similar manner or whether mtDNA is essential for tumorigenesis. We show that human neural stem cells (hNSCs) increased their mtDNA content during differentiation in a process that was mediated by a synergistic relationship between the nuclear and mitochondrial genomes and results in increased respiratory capacity. Differentiating multipotent glioblastoma cells failed to match the expansion in mtDNA copy number, patterns of gene expression and increased respiratory capacity observed in hNSCs. Partial depletion of glioblastoma cell mtDNA rescued mtDNA replication events and enhanced cell differentiation. However, prolonged depletion resulted in impaired mtDNA replication, reduced proliferation and induced the expression of early developmental and pro-survival markers including POU class 5 homeobox 1 (OCT4) and sonic hedgehog (SHH). The transfer of glioblastoma cells depleted to varying degrees of their mtDNA content into immunocompromised mice resulted in tumors requiring significantly longer to form compared with non-depleted cells. The number of tumors formed and the time to tumor formation was relative to the degree of mtDNA depletion. The tumors derived from mtDNA depleted glioblastoma cells recovered their mtDNA copy number as part of the tumor formation process. These outcomes demonstrate the importance of mtDNA to the initiation and maintenance of tumorigenesis in glioblastoma multiforme.
Figures




Comment in
-
When numbers matters: mitochondrial DNA and gliomagenesis.Cell Death Differ. 2013 Dec;20(12):1601-2. doi: 10.1038/cdd.2013.156. Cell Death Differ. 2013. PMID: 24212930 Free PMC article. No abstract available.
References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. - PubMed
-
- Clayton DA. Nuclear-mitochondrial intergenomic communication. BioFactors. 1998;7:203–205. - PubMed
-
- Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. - PubMed
-
- Kucej M, Butow RA. Evolutionary tinkering with mitochondrial nucleoids. Trends Cell Biol. 2007;17:586–592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

