Mechanisms of tumor cell extravasation in an in vitro microvascular network platform
- PMID: 23995847
- PMCID: PMC4038741
- DOI: 10.1039/c3ib40149a
Mechanisms of tumor cell extravasation in an in vitro microvascular network platform
Abstract
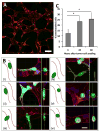
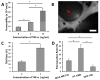
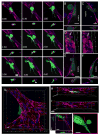
A deeper understanding of the mechanisms of tumor cell extravasation is essential in creating therapies that target this crucial step in cancer metastasis. Here, we use a microfluidic platform to study tumor cell extravasation from in vitro microvascular networks formed via vasculogenesis. We demonstrate tight endothelial cell-cell junctions, basement membrane deposition and physiological values of vessel permeability. Employing our assay, we demonstrate impaired endothelial barrier function and increased extravasation efficiency with inflammatory cytokine stimulation, as well as positive correlations between the metastatic potentials of MDA-MB-231, HT-1080, MCF-10A and their extravasation capabilities. High-resolution time-lapse microscopy reveals the highly dynamic nature of extravasation events, beginning with thin tumor cell protrusions across the endothelium followed by extrusion of the remainder of the cell body through the formation of small (~1 μm) openings in the endothelial barrier which grows in size (~8 μm) to allow for nuclear transmigration. No disruption to endothelial cell-cell junctions is discernible at 60×, or by changes in local barrier function after completion of transmigration. Tumor transendothelial migration efficiency is significantly higher in trapped cells compared to non-trapped adhered cells, and in cell clusters versus single tumor cells.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

