Functional genotyping of Sulfurospirillum spp. in mixed cultures allowed the identification of a new tetrachloroethene reductive dehalogenase
- PMID: 23995945
- PMCID: PMC3811552
- DOI: 10.1128/AEM.02312-13
Functional genotyping of Sulfurospirillum spp. in mixed cultures allowed the identification of a new tetrachloroethene reductive dehalogenase
Abstract
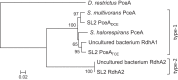
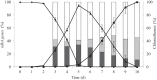
Reductive dehalogenases are the key enzymes involved in the anaerobic respiration of organohalides such as the widespread groundwater pollutant tetrachloroethene. The increasing number of available bacterial genomes and metagenomes gives access to hundreds of new putative reductive dehalogenase genes that display a high level of sequence diversity and for which substrate prediction remains very challenging. In this study, we present the development of a functional genotyping method targeting the diverse reductive dehalogenases present in Sulfurospirillum spp., which allowed us to unambiguously identify a new reductive dehalogenase from our tetrachloroethene-dechlorinating SL2 bacterial consortia. The new enzyme, named PceATCE, shows 92% sequence identity with the well-characterized PceA enzyme of Sulfurospirillum multivorans, but in contrast to the latter, it is restricted to tetrachloroethene as a substrate. Its apparent higher dechlorinating activity with tetrachloroethene likely allowed its selection and maintenance in the bacterial consortia among other enzymes showing broader substrate ranges. The sequence-substrate relationships within tetrachloroethene reductive dehalogenases are also discussed.
Figures

References
-
- Villemur R, Lanthier M, Beaudet R, Lepine F. 2006. The Desulfitobacterium genus. FEMS Microbiol. Rev. 30:706–733 - PubMed
-
- Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Muller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63:625–635 - PubMed
-
- Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJ. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313–321 - PubMed
-
- Scholz-Muramatsu H, Neumann A, Messmer M, Moore E, Diekert G. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163:48–56
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

