Biophysical basis of the promiscuous binding of B-cell lymphoma protein 2 apoptotic repressor to BH3 ligands
- PMID: 23996493
- PMCID: PMC3761409
- DOI: 10.1002/jmr.2295
Biophysical basis of the promiscuous binding of B-cell lymphoma protein 2 apoptotic repressor to BH3 ligands
Abstract
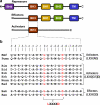
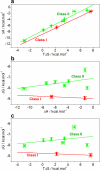
B-cell lymphoma protein 2 (Bcl2) apoptotic repressor carries out its function by virtue of its ability to bind to BH3 domains of various pro-apoptotic regulators in a highly promiscuous manner. Herein, we investigate the biophysical basis of such promiscuity of Bcl2 toward its cognate BH3 ligands. Our data show that although the BH3 ligands harboring the LXXXAD motif bind to Bcl2 with submicromolar affinity, those with the LXXX[G/S]D motif afford weak interactions. This implies that the replacement of alanine at the fourth position (A + 4)-relative to the N-terminal leucine (L0) within the LXXXAD motif-to glycine/serine results in the loss of free energy of binding. Consistent with this notion, the A + 4 residue within the BH3 ligands harboring the LXXXAD motif engages in key intermolecular van der Waals contacts with A149 lining the ligand binding groove within Bcl2, whereas A + 4G/S substitution results in the disruption of such favorable binding interactions. Of particular interest is the observation that although increasing ionic strength has little or negligible effect on the binding of high-affinity BH3 ligands harboring the LXXXAD motif, the binding of those with the LXXX[G/S]D motif in general experiences a varying degree of enhancement. This salient observation is indicative of the fact that hydrophobic forces not only play a dominant but also a universal role in driving the Bcl2-BH3 interactions. Taken together, our study sheds light on the molecular basis of the factors governing the promiscuous binding of Bcl2 to pro-apoptotic regulators and thus bears important consequences on the development of rational therapeutic approaches.
Keywords: binding thermodynamics; molecular dynamics; salt dependence; structural models.
Copyright © 2013 John Wiley & Sons, Ltd.
Figures






References
-
- Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. - PubMed
-
- Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J Phys Chem. 1987;91:6269–6271.
-
- Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

