Nkx2.2:Cre knock-in mouse line: a novel tool for pancreas- and CNS-specific gene deletion
- PMID: 23996959
- PMCID: PMC3902634
- DOI: 10.1002/dvg.22715
Nkx2.2:Cre knock-in mouse line: a novel tool for pancreas- and CNS-specific gene deletion
Abstract
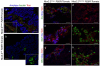
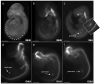
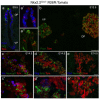
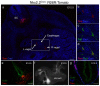
Nkx2.2 is a homeodomain-containing transcriptional regulator necessary for the appropriate differentiation of ventral neuronal populations in the spinal cord and hindbrain, and endocrine cell populations in the pancreas and intestine. In each tissue, Nkx2.2 inactivation leads to reciprocal cell fate alterations. To confirm the cell fate changes are due to respecification of Nkx2.2-expressing progenitors and to provide a novel tool for lineage tracing in the pancreas and CNS, we generated an Nkx2.2:Cre mouse line by knocking in a Cre-EGFP cassette into the Nkx2.2 genomic locus and inactivating endogenous Nkx2.2. The R26R-CAG-LSL-tdTomato reporter was used to monitor the specificity and efficiency of Nkx2.2:Cre activity; the tomato reporter faithfully recapitulated endogenous Nkx2.2 expression and could be detected as early as embryonic day (e) 9.25 in the developing CNS and was initiated shortly thereafter at e9.5 in the pancreas. Lineage analyses in the CNS confirmed the cell populations thought to be derived from Nkx2.2-expressing progenitor domains. Furthermore, lineage studies verified Nkx2.2 expression in the earliest pancreatic progenitors that give rise to all cell types of the pancreas; however they also revealed more robust Cre activity in the dorsal versus ventral pancreas. Thus, the Nkx2.2:Cre line provides a novel tool for gene manipulations in the CNS and pancreas.
Keywords: CNS; Cre-lox; Nkx2.2; lineage; pancreas.
Copyright © 2013 Wiley Periodicals, Inc.
Figures


Similar articles
-
Generation of a Nkx2.2(Cre) knock-in mouse line: Analysis of cell lineages in the central nervous system.Differentiation. 2015 Mar-Apr;89(3-4):70-6. doi: 10.1016/j.diff.2015.03.001. Epub 2015 Apr 1. Differentiation. 2015. PMID: 25840610
-
Generation of Nkx2.2:lacZ mice using recombination-mediated cassette exchange technology.Genesis. 2012 Aug;50(8):612-24. doi: 10.1002/dvg.22037. Epub 2012 May 19. Genesis. 2012. PMID: 22539496 Free PMC article.
-
Generation of mice encoding a conditional allele of Nkx2.2.Transgenic Res. 2013 Oct;22(5):965-72. doi: 10.1007/s11248-013-9700-0. Epub 2013 Mar 15. Transgenic Res. 2013. PMID: 23494546 Free PMC article.
-
Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5.Int J Dev Biol. 2002;46(4):431-9. Int J Dev Biol. 2002. PMID: 12141429
-
Differential use of the Nkx2.2 NK2 domain in developing pancreatic islets and neurons.Genes Dev. 2023 Jun 1;37(11-12):451-453. doi: 10.1101/gad.350895.123. Epub 2023 Jun 30. Genes Dev. 2023. PMID: 37399332 Free PMC article. Review.
Cited by
-
Corticospinal Circuits from the Sensory and Motor Cortices Differentially Regulate Skilled Movements through Distinct Spinal Interneurons.Cell Rep. 2018 May 1;23(5):1286-1300.e7. doi: 10.1016/j.celrep.2018.03.137. Cell Rep. 2018. PMID: 29719245 Free PMC article.
-
From classical to current: analyzing peripheral nervous system and spinal cord lineage and fate.Dev Biol. 2015 Feb 15;398(2):135-46. doi: 10.1016/j.ydbio.2014.09.033. Epub 2014 Oct 24. Dev Biol. 2015. PMID: 25446276 Free PMC article. Review.
-
Overexpression of Truncated Human DISC1 Induces Appearance of Hindbrain Oligodendroglia in the Forebrain During Development.Schizophr Bull. 2018 Apr 6;44(3):515-524. doi: 10.1093/schbul/sbx106. Schizophr Bull. 2018. PMID: 28981898 Free PMC article.
-
Decoding gene networks controlling hypothalamic and prethalamic neuron development.Cell Rep. 2025 Jun 24;44(6):115858. doi: 10.1016/j.celrep.2025.115858. Epub 2025 Jun 12. Cell Rep. 2025. PMID: 40512619 Free PMC article.
-
Major β cell-specific functions of NKX2.2 are mediated via the NK2-specific domain.Genes Dev. 2023 Jun 1;37(11-12):490-504. doi: 10.1101/gad.350569.123. Epub 2023 Jun 26. Genes Dev. 2023. PMID: 37364986 Free PMC article.
References
-
- Briscoe J, Sussel L, Serup P, Hartigan-O’Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeo-box gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. - PubMed
-
- Chen SX, Osipovich AB, Ustione A, Potter LA, Hipkens S, Gangula R, Yuan W, Piston DW, Magnuson MA. Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis Model Mech. 2011 DOI:dmm.006569 [pii] 10.1242/dmm.006569. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

