Common DNA methylation alterations in multiple brain regions in autism
- PMID: 23999529
- PMCID: PMC4184909
- DOI: 10.1038/mp.2013.114
Common DNA methylation alterations in multiple brain regions in autism
Abstract
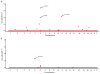
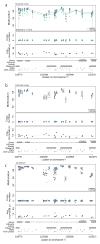
Autism spectrum disorders (ASD) are increasingly common neurodevelopmental disorders defined clinically by a triad of features including impairment in social interaction, impairment in communication in social situations and restricted and repetitive patterns of behavior and interests, with considerable phenotypic heterogeneity among individuals. Although heritability estimates for ASD are high, conventional genetic-based efforts to identify genes involved in ASD have yielded only few reproducible candidate genes that account for only a small proportion of ASDs. There is mounting evidence to suggest environmental and epigenetic factors play a stronger role in the etiology of ASD than previously thought. To begin to understand the contribution of epigenetics to ASD, we have examined DNA methylation (DNAm) in a pilot study of postmortem brain tissue from 19 autism cases and 21 unrelated controls, among three brain regions including dorsolateral prefrontal cortex, temporal cortex and cerebellum. We measured over 485,000 CpG loci across a diverse set of functionally relevant genomic regions using the Infinium HumanMethylation450 BeadChip and identified four genome-wide significant differentially methylated regions (DMRs) using a bump hunting approach and a permutation-based multiple testing correction method. We replicated 3/4 DMRs identified in our genome-wide screen in a different set of samples and across different brain regions. The DMRs identified in this study represent suggestive evidence for commonly altered methylation sites in ASD and provide several promising new candidate genes.
Conflict of interest statement
The authors declare no conflict of interest
Figures


References
-
- Rapin I. Autism. N Engl J Med. 1997;337(2):97–104. - PubMed
-
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4. American Psychiatric Association Press; 1994.
-
- Autism: economic impact and implications. Proceedings of the 2012 Autism Summit; March 31, 2012; Hong Kong, China. 2012.
-
- Dawson G. Dramatic Increase in Autism Prevalence Parallels Explosion of Research Into Its Biology and Causes. Arch Gen Psychiatry. 2012:1–2. - PubMed
-
- Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT. CACNA1H mutations in autism spectrum disorders. J Biol Chem. 2006;281(31):22085–22091. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

