Evaluating the impact of pre-annotation on annotation speed and potential bias: natural language processing gold standard development for clinical named entity recognition in clinical trial announcements
- PMID: 24001514
- PMCID: PMC3994857
- DOI: 10.1136/amiajnl-2013-001837
Evaluating the impact of pre-annotation on annotation speed and potential bias: natural language processing gold standard development for clinical named entity recognition in clinical trial announcements
Abstract
Objective: To present a series of experiments: (1) to evaluate the impact of pre-annotation on the speed of manual annotation of clinical trial announcements; and (2) to test for potential bias, if pre-annotation is utilized.
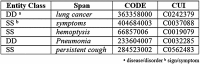
Methods: To build the gold standard, 1400 clinical trial announcements from the clinicaltrials.gov website were randomly selected and double annotated for diagnoses, signs, symptoms, Unified Medical Language System (UMLS) Concept Unique Identifiers, and SNOMED CT codes. We used two dictionary-based methods to pre-annotate the text. We evaluated the annotation time and potential bias through F-measures and ANOVA tests and implemented Bonferroni correction.
Results: Time savings ranged from 13.85% to 21.5% per entity. Inter-annotator agreement (IAA) ranged from 93.4% to 95.5%. There was no statistically significant difference for IAA and annotator performance in pre-annotations.
Conclusions: On every experiment pair, the annotator with the pre-annotated text needed less time to annotate than the annotator with non-labeled text. The time savings were statistically significant. Moreover, the pre-annotation did not reduce the IAA or annotator performance. Dictionary-based pre-annotation is a feasible and practical method to reduce the cost of annotation of clinical named entity recognition in the eligibility sections of clinical trial announcements without introducing bias in the annotation process.
Keywords: Information Extraction; Natural Language Processing; Pre-annotation; clinical trial announcements; named entity recognition; umls.
Figures
Similar articles
-
Web 2.0-based crowdsourcing for high-quality gold standard development in clinical natural language processing.J Med Internet Res. 2013 Apr 2;15(4):e73. doi: 10.2196/jmir.2426. J Med Internet Res. 2013. PMID: 23548263 Free PMC article.
-
A multilingual gold-standard corpus for biomedical concept recognition: the Mantra GSC.J Am Med Inform Assoc. 2015 Sep;22(5):948-56. doi: 10.1093/jamia/ocv037. Epub 2015 May 6. J Am Med Inform Assoc. 2015. PMID: 25948699 Free PMC article.
-
A clinical trials corpus annotated with UMLS entities to enhance the access to evidence-based medicine.BMC Med Inform Decis Mak. 2021 Feb 22;21(1):69. doi: 10.1186/s12911-021-01395-z. BMC Med Inform Decis Mak. 2021. PMID: 33618727 Free PMC article.
-
Accelerating the annotation of sparse named entities by dynamic sentence selection.BMC Bioinformatics. 2008 Nov 19;9 Suppl 11(Suppl 11):S8. doi: 10.1186/1471-2105-9-S11-S8. BMC Bioinformatics. 2008. PMID: 19025694 Free PMC article.
-
Quantitative analysis of manual annotation of clinical text samples.Int J Med Inform. 2019 Mar;123:37-48. doi: 10.1016/j.ijmedinf.2018.12.011. Epub 2018 Dec 31. Int J Med Inform. 2019. PMID: 30654902
Cited by
-
Clinical Natural Language Processing in 2014: Foundational Methods Supporting Efficient Healthcare.Yearb Med Inform. 2015 Aug 13;10(1):194-8. doi: 10.15265/IY-2015-035. Yearb Med Inform. 2015. PMID: 26293868 Free PMC article.
-
Adverse drug event detection using natural language processing: A scoping review of supervised learning methods.PLoS One. 2023 Jan 3;18(1):e0279842. doi: 10.1371/journal.pone.0279842. eCollection 2023. PLoS One. 2023. PMID: 36595517 Free PMC article.
-
ADE Eval: An Evaluation of Text Processing Systems for Adverse Event Extraction from Drug Labels for Pharmacovigilance.Drug Saf. 2021 Jan;44(1):83-94. doi: 10.1007/s40264-020-00996-3. Epub 2020 Oct 2. Drug Saf. 2021. PMID: 33006728 Free PMC article.
-
Identification of social determinants of health using multi-label classification of electronic health record clinical notes.JAMIA Open. 2021 Feb 9;4(3):ooaa069. doi: 10.1093/jamiaopen/ooaa069. eCollection 2021 Jul. JAMIA Open. 2021. PMID: 34514351 Free PMC article.
-
Using Nonexperts for Annotating Pharmacokinetic Drug-Drug Interaction Mentions in Product Labeling: A Feasibility Study.JMIR Res Protoc. 2016 Apr 11;5(2):e40. doi: 10.2196/resprot.5028. JMIR Res Protoc. 2016. PMID: 27066806 Free PMC article.
References
-
- Tomanek K, Wermter J, Hahn U. Efficient annotation with the jena annotation environment (JANE). Linguistic Annotation Workshop—A Merger of NLPXML 2007 and FLAC 2007; Prague, Czech Republic: Association for Computational Linguistics (ACL), 2007:9–16
-
- Ganchev K, Pereira F, Mandel M, et al. Semi-automated named entity annotation. Proceedings of the Linguistic Annotation Workshop; Prague, Czech Republic: Association for Computational Linguistics, 2007:53–6
-
- Aramaki E, Miura Y, Tonoike M, et al. Extraction of adverse drug effects from clinical records. Stud Health Technol Inform 2010;160(Pt 1):739–43 - PubMed
-
- Ogren P, Savova G, Chute C. Constructing evaluation corpora for automated clinical named entity recognition. Proceedings of the Language Resources and Evaluation Conference (LREC); 2008:28–30
-
- Hachey B, Alex B, Becker M. Investigating the effects of selective sampling on the annotation task. Proceedings of the Ninth Conference on Computational Natural Language Learning; Ann Arbor, Michigan: Association for Computational Linguistics, 2005:144–51
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical