Evaluating the impact of pre-annotation on annotation speed and potential bias: natural language processing gold standard development for clinical named entity recognition in clinical trial announcements
- PMID: 24001514
- PMCID: PMC3994857
- DOI: 10.1136/amiajnl-2013-001837
Evaluating the impact of pre-annotation on annotation speed and potential bias: natural language processing gold standard development for clinical named entity recognition in clinical trial announcements
Abstract
Objective: To present a series of experiments: (1) to evaluate the impact of pre-annotation on the speed of manual annotation of clinical trial announcements; and (2) to test for potential bias, if pre-annotation is utilized.
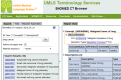
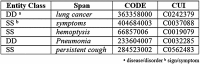
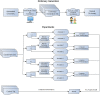
Methods: To build the gold standard, 1400 clinical trial announcements from the clinicaltrials.gov website were randomly selected and double annotated for diagnoses, signs, symptoms, Unified Medical Language System (UMLS) Concept Unique Identifiers, and SNOMED CT codes. We used two dictionary-based methods to pre-annotate the text. We evaluated the annotation time and potential bias through F-measures and ANOVA tests and implemented Bonferroni correction.
Results: Time savings ranged from 13.85% to 21.5% per entity. Inter-annotator agreement (IAA) ranged from 93.4% to 95.5%. There was no statistically significant difference for IAA and annotator performance in pre-annotations.
Conclusions: On every experiment pair, the annotator with the pre-annotated text needed less time to annotate than the annotator with non-labeled text. The time savings were statistically significant. Moreover, the pre-annotation did not reduce the IAA or annotator performance. Dictionary-based pre-annotation is a feasible and practical method to reduce the cost of annotation of clinical named entity recognition in the eligibility sections of clinical trial announcements without introducing bias in the annotation process.
Keywords: Information Extraction; Natural Language Processing; Pre-annotation; clinical trial announcements; named entity recognition; umls.
Figures
References
-
- Tomanek K, Wermter J, Hahn U. Efficient annotation with the jena annotation environment (JANE). Linguistic Annotation Workshop—A Merger of NLPXML 2007 and FLAC 2007; Prague, Czech Republic: Association for Computational Linguistics (ACL), 2007:9–16
-
- Ganchev K, Pereira F, Mandel M, et al. Semi-automated named entity annotation. Proceedings of the Linguistic Annotation Workshop; Prague, Czech Republic: Association for Computational Linguistics, 2007:53–6
-
- Aramaki E, Miura Y, Tonoike M, et al. Extraction of adverse drug effects from clinical records. Stud Health Technol Inform 2010;160(Pt 1):739–43 - PubMed
-
- Ogren P, Savova G, Chute C. Constructing evaluation corpora for automated clinical named entity recognition. Proceedings of the Language Resources and Evaluation Conference (LREC); 2008:28–30
-
- Hachey B, Alex B, Becker M. Investigating the effects of selective sampling on the annotation task. Proceedings of the Ninth Conference on Computational Natural Language Learning; Ann Arbor, Michigan: Association for Computational Linguistics, 2005:144–51
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical