RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core
- PMID: 24002212
- PMCID: PMC3817461
- DOI: 10.1038/emboj.2013.198
RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core
Abstract
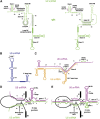
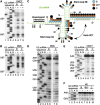
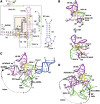
Although U snRNAs play essential roles in splicing, little is known about the 3D arrangement of U2, U6, and U5 snRNAs and the pre-mRNA in active spliceosomes. To elucidate their relative spatial organization and dynamic rearrangement, we examined the RNA structure of affinity-purified, human spliceosomes before and after catalytic step 1 by chemical RNA structure probing. We found a stable 3-way junction of the U2/U6 snRNA duplex in active spliceosomes that persists minimally through step 1. Moreover, the formation of alternating, mutually exclusive, U2 snRNA conformations, as observed in yeast, was not detected in different assembly stages of human spliceosomal complexes (that is, B, B(act), or C complexes). Psoralen crosslinking revealed an interaction during/after step 1 between internal loop 1 of the U5 snRNA, and intron nucleotides immediately downstream of the branchpoint. Using the experimentally derived structural constraints, we generated a model of the RNA network of the step 1 spliceosome, based on the crystal structure of a group II intron through homology modelling. The model is topologically consistent with current genetic, biochemical, and structural data.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






Comment in
-
RNAtomy of the Spliceosome's heart.EMBO J. 2013 Oct 30;32(21):2785-7. doi: 10.1038/emboj.2013.213. Epub 2013 Sep 24. EMBO J. 2013. PMID: 24065126 Free PMC article.
References
-
- Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Lührmann R (2010) Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16: 2384–2403 - PMC - PubMed
-
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R (2008) Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

