Nucleosome remodeling and epigenetics
- PMID: 24003213
- PMCID: PMC3753709
- DOI: 10.1101/cshperspect.a017905
Nucleosome remodeling and epigenetics
Abstract
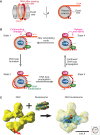
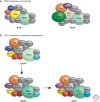
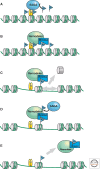
Eukaryotic chromatin is kept flexible and dynamic to respond to environmental, metabolic, and developmental cues through the action of a family of so-called "nucleosome remodeling" ATPases. Consistent with their helicase ancestry, these enzymes experience conformation changes as they bind and hydrolyze ATP. At the same time they interact with DNA and histones, which alters histone-DNA interactions in target nucleosomes. Their action may lead to complete or partial disassembly of nucleosomes, the exchange of histones for variants, the assembly of nucleosomes, or the movement of histone octamers on DNA. "Remodeling" may render DNA sequences accessible to interacting proteins or, conversely, promote packing into tightly folded structures. Remodeling processes participate in every aspect of genome function. Remodeling activities are commonly integrated with other mechanisms such as histone modifications or RNA metabolism to assemble stable, epigenetic states.
Figures






References
-
- Altaf M, Auger A, Covic M, Cote J 2009. Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol 87: 35–50 - PubMed
-
- Altaf M, Saksouk N, Cote J 2007. Histone modifications in response to DNA damage. Mutat Res 618: 81–90 - PubMed
-
- Bao Y, Shen X 2007. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev 17: 126–131 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
