RNA-mediated regulation in pathogenic bacteria
- PMID: 24003243
- PMCID: PMC3753719
- DOI: 10.1101/cshperspect.a010298
RNA-mediated regulation in pathogenic bacteria
Abstract
Pathogenic bacteria possess intricate regulatory networks that temporally control the production of virulence factors, and enable the bacteria to survive and proliferate after host infection. Regulatory RNAs are now recognized as important components of these networks, and their study may not only identify new approaches to combat infectious diseases but also reveal new general control mechanisms involved in bacterial gene expression. In this review, we illustrate the diversity of regulatory RNAs in bacterial pathogens, their mechanism of action, and how they can be integrated into the regulatory circuits that govern virulence-factor production.
Figures



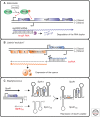

References
-
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 - PubMed
-
- Balaban N, Novick RP 1995. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′ end deletion. FEMS Microbiol Lett 133: 155–161 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
