BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state
- PMID: 24004950
- PMCID: PMC3775414
- DOI: 10.1242/dev.092155
BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state
Abstract
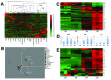
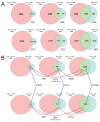
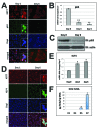
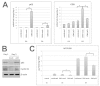
The placenta is a transient organ that is necessary for proper fetal development. Its main functional component is the trophoblast, which is derived from extra-embryonic ectoderm. Little is known about early trophoblast differentiation in the human embryo, owing to lack of a proper in vitro model system. Human embryonic stem cells (hESCs) differentiate into functional trophoblast following BMP4 treatment in the presence of feeder-conditioned media; however, this model has not been widely accepted, in part owing to a lack of proof for a trophoblast progenitor population. We have previously shown that p63, a member of the p53 family of nuclear proteins, is expressed in proliferative cytotrophoblast (CTB), precursors to terminally differentiated syncytiotrophoblast (STB) in chorionic villi and extravillous trophoblast (EVT) at the implantation site. Here, we show that BMP4-treated hESCs differentiate into bona fide CTB by direct comparison with primary human placental tissues and isolated CTB through gene expression profiling. We show that, in primary CTB, p63 levels are reduced as cells differentiate into STB, and that forced expression of p63 maintains cyclin B1 and inhibits STB differentiation. We also establish that, similar to in vivo events, hESC differentiation into trophoblast is characterized by a p63(+)/KRT7(+) CTB stem cell state, followed by formation of functional KLF4(+) STB and HLA-G(+) EVT. Finally, we illustrate that downregulation of p63 by shRNA inhibits differentiation of hESCs into functional trophoblast. Taken together, our results establish that BMP4-treated hESCs are an excellent model of human trophoblast differentiation, closely mimicking the in vivo progression from p63(+) CTB stem cells to terminally differentiated trophoblast subtypes.
Keywords: BMP4; Human embryonic stem cells; Placenta; Trophoblast; ΔNp63.
Figures


References
-
- Adjaye J., Huntriss J., Herwig R., BenKahla A., Brink T. C., Wierling C., Hultschig C., Groth D., Yaspo M. L., Picton H. M., et al. (2005). Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells 23, 1514–1525 - PubMed
-
- Apps R., Murphy S. P., Fernando R., Gardner L., Ahad T., Moffett A. (2009). Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology 127, 26–39 - PMC - PubMed
-
- Baczyk D., Drewlo S., Proctor L., Dunk C., Lye S., Kingdom J. (2009). Glial cell missing-1 transcription factor is required for the differentiation of the human trophoblast. Cell Death Differ. 16, 719–727 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

