Effects of sleep and wake on oligodendrocytes and their precursors
- PMID: 24005282
- PMCID: PMC3874087
- DOI: 10.1523/JNEUROSCI.5102-12.2013
Effects of sleep and wake on oligodendrocytes and their precursors
Abstract
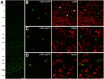
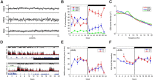
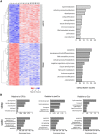
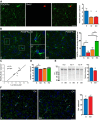
Previous studies of differential gene expression in sleep and wake pooled transcripts from all brain cells and showed that several genes expressed at higher levels during sleep are involved in the synthesis/maintenance of membranes in general and of myelin in particular, a surprising finding given the reported slow turnover of many myelin components. Other studies showed that oligodendrocyte precursor cells (OPCs) are responsible for the formation of new myelin in both the injured and the normal adult brain, and that glutamate released from neurons, via neuron-OPC synapses, can inhibit OPC proliferation and affect their differentiation into myelin-forming oligodendrocytes. Because glutamatergic transmission is higher in wake than in sleep, we asked whether sleep and wake can affect oligodendrocytes and OPCs. Using the translating ribosome affinity purification technology combined with microarray analysis in mice, we obtained a genome-wide profiling of oligodendrocytes after sleep, spontaneous wake, and forced wake (acute sleep deprivation). We found that hundreds of transcripts being translated in oligodendrocytes are differentially expressed in sleep and wake: genes involved in phospholipid synthesis and myelination or promoting OPC proliferation are transcribed preferentially during sleep, while genes implicated in apoptosis, cellular stress response, and OPC differentiation are enriched in wake. We then confirmed through BrdU and other experiments that OPC proliferation doubles during sleep and positively correlates with time spent in REM sleep, whereas OPC differentiation is higher during wake. Thus, OPC proliferation and differentiation are not perfectly matched at any given circadian time but preferentially occur during sleep and wake, respectively.
Figures

References
-
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
