Identification of amino acids conferring chain length substrate specificities on fatty alcohol-forming reductases FAR5 and FAR8 from Arabidopsis thaliana
- PMID: 24005667
- PMCID: PMC3798499
- DOI: 10.1074/jbc.M113.499715
Identification of amino acids conferring chain length substrate specificities on fatty alcohol-forming reductases FAR5 and FAR8 from Arabidopsis thaliana
Abstract
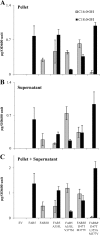
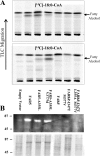
Fatty alcohols play a variety of biological roles in all kingdoms of life. Fatty acyl reductase (FAR) enzymes catalyze the reduction of fatty acyl-coenzyme A (CoA) or fatty acyl-acyl carrier protein substrates to primary fatty alcohols. FAR enzymes have distinct substrate specificities with regard to chain length and degree of saturation. FAR5 (At3g44550) and FAR8 (At3g44560) from Arabidopsis thaliana are 85% identical at the amino acid level and are of equal length, but they possess distinct specificities for 18:0 or 16:0 acyl chain length, respectively. We used Saccharomyces cerevisiae as a heterologous expression system to assess FAR substrate specificity determinants. We identified individual amino acids that affect protein levels or 16:0-CoA versus 18:0-CoA specificity by expressing in yeast FAR5 and FAR8 domain-swap chimeras and site-specific mutants. We found that a threonine at position 347 and a serine at position 363 were important for high FAR5 and FAR8 protein accumulation in yeast and thus are likely important for protein folding and stability. Amino acids at positions 355 and 377 were important for dictating 16:0-CoA versus 18:0-CoA chain length specificity. Simultaneously converting alanine 355 and valine 377 of FAR5 to the corresponding FAR8 residues, leucine and methionine, respectively, almost fully converted FAR5 specificity from 18:0-CoA to 16:0-CoA. The reciprocal amino acid conversions, L355A and M377V, made in the active FAR8-S363P mutant background converted its specificity from 16:0-CoA to 18:0-CoA. This study is an important advancement in the engineering of highly active FAR proteins with desired specificities for the production of fatty alcohols with industrial value.
Keywords: Enzyme Structure; Fatty Acyl Reductase; Fatty Alcohol; Lipid Metabolism; Plant Biochemistry; Protein Engineering; Site-directed Mutagenesis.
Figures



References
-
- Rowland O., Domergue F. (2012) Plant fatty acyl reductases: enzymes generating fatty alcohols for protective layers with potential for industrial applications. Plant Sci. 193, 28–38 - PubMed
-
- Antia N. J., Lee R. F., Nevenzel J. C., Cheng J. Y. (1974) Wax ester production by the marine cryptomonad Chroomonas salina grown photoheterotrophically on glycerol. J. Protozool. 21, 768–771 - PubMed
-
- Teerawanichpan P., Robertson A. J., Qiu X. (2010) A fatty acyl-CoA reductase highly expressed in the head of honey bee (Apis mellifera) involves biosynthesis of a wide range of aliphatic fatty alcohols. Insect Biochem. Mol. Biol. 40, 641–649 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

