Inside-out signaling promotes dynamic changes in the carcinoembryonic antigen-related cellular adhesion molecule 1 (CEACAM1) oligomeric state to control its cell adhesion properties
- PMID: 24005674
- PMCID: PMC3795263
- DOI: 10.1074/jbc.M113.504639
Inside-out signaling promotes dynamic changes in the carcinoembryonic antigen-related cellular adhesion molecule 1 (CEACAM1) oligomeric state to control its cell adhesion properties
Abstract
Cell-cell contacts are fundamental to multicellular organisms and are subject to exquisite levels of control. The carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) can engage in both cis-homophilic (parallel) oligomerization and trans-homophilic (anti-parallel) binding. In this study, we establish that the CEACAM1 transmembrane domain has a propensity to form cis-dimers via the transmembrane-embedded (432)GXXXG(436) motif and that this basal state is overcome when activated calmodulin binds to the CEACAM1 cytoplasmic domain. Although mutation of the (432)GXXXG(436) motif reduced CEACAM1 oligomerization, it did not affect surface localization of the receptor or influence CEACAM1-dependent cellular invasion by the pathogenic Neisseria. The mutation did, however, have a striking effect on CEACAM1-dependent cellular aggregation, increasing both the kinetics of cell-cell association and the size of cellular aggregates formed. CEACAM1 association with tyrosine kinase c-Src and tyrosine phosphatases SHP-1 and SHP-2 was not affected by the (432)GXXXG(436) mutation, consistent with their association with the monomeric form of wild type CEACAM1. Collectively, our results establish that a dynamic oligomer-to-monomer shift in surface-expressed CEACAM1 facilitates trans-homophilic binding and downstream effector signaling.
Keywords: Bacterial Adhesion; CEACAM1; Calcium Signaling; Calmodulin; Cancer; Carcinoembryonic Antigen-related Cell Adhesion Molecule; Cell Adhesion; Homodimer; Homophilic Binding; Membrane Proteins.
Figures

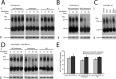
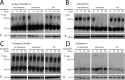

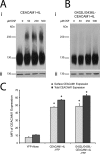
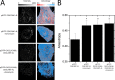

References
-
- Okegawa T., Pong R. C., Li Y., Hsieh J. T. (2004) The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim. Pol. 51, 445–457 - PubMed
-
- Beauchemin N., Draber P., Dveksler G., Gold P., Gray-Owen S., Grunert F., Hammarström S., Holmes K. V., Karlsson A., Kuroki M., Lin S. H., Lucka L., Najjar S. M., Neumaier M., Obrink B., Shively J. E., Skubitz K. M., Stanners C. P., Thomas P., Thompson J. A., Virji M., von Kleist S., Wagener C., Watt S., Zimmermann W. (1999) Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252, 243–249 - PubMed
-
- Hinoda Y., Neumaier M., Hefta S. A., Drzeniek Z., Wagener C., Shively L., Hefta L. J., Shively J. E., Paxton R. J. (1988) Molecular cloning of a cDNA coding biliary glycoprotein I: primary structure of a glycoprotein immunologically cross-reactive with carcinoembryonic antigen. Proc. Natl. Acad. Sci. U.S.A. 85, 6959–6963 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

