Increased expression of reticulon 3 in neurons leads to reduced axonal transport of β site amyloid precursor protein-cleaving enzyme 1
- PMID: 24005676
- PMCID: PMC3798490
- DOI: 10.1074/jbc.M113.480079
Increased expression of reticulon 3 in neurons leads to reduced axonal transport of β site amyloid precursor protein-cleaving enzyme 1
Abstract
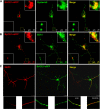
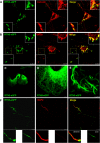
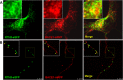
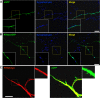
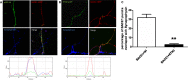
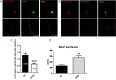
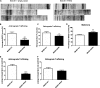
BACE1 is the sole enzyme responsible for cleaving amyloid precursor protein at the β-secretase site, and this cleavage initiates the generation of β-amyloid peptide (Aβ). Because amyloid precursor protein is predominantly expressed by neurons and deposition of Aβ aggregates in the human brain is highly correlated with the Aβ released at axonal terminals, we focused our investigation of BACE1 localization on the neuritic region. We show that BACE1 was not only enriched in the late Golgi, trans-Golgi network, and early endosomes but also in both axons and dendrites. BACE1 was colocalized with the presynaptic vesicle marker synaptophysin, indicating the presence of BACE1 in synapses. Because the excessive release of Aβ from synapses is attributable to an increase in amyloid deposition, we further explored whether the presence of BACE1 in synapses was regulated by reticulon 3 (RTN3), a protein identified previously as a negative regulator of BACE1. We found that RTN3 is not only localized in the endoplasmic reticulum but also in neuritic regions where no endoplasmic reticulum-shaping proteins are detected, implicating additional functions of RTN3 in neurons. Coexpression of RTN3 with BACE1 in cultured neurons was sufficient to reduce colocalization of BACE1 with synaptophysin. This reduction correlated with decreased anterograde transport of BACE1 in axons in response to overexpressed RTN3. Our results in this study suggest that altered RTN3 levels can impact the axonal transport of BACE1 and demonstrate that reducing axonal transport of BACE1 in axons is a viable strategy for decreasing BACE1 in axonal terminals and, perhaps, reducing amyloid deposition.
Keywords: Alzheimer Disease; Amyloid Precursor Protein; Axonal Transport; BACE1; Confocal Microscopy; Kymograph; RTN3; Secretases; Synapses; Synaptic Localization.
Figures


References
-
- Selkoe D. J., Yamazaki T., Citron M., Podlisny M. B., Koo E. H., Teplow D. B., Haass C. (1996) The role of APP processing and trafficking pathways in the formation of amyloid β-protein. Ann. N.Y. Acad. Sci. 777, 57–64 - PubMed
-
- Sisodia S. S., St George-Hyslop P. H. (2002) γ-Secretase, Notch, Aβ and Alzheimer's disease. Where do the presenilins fit in? Nat. Rev. Neurosci. 3, 281–290 - PubMed
-
- Hussain I., Powell D., Howlett D. R., Tew D. G., Meek T. D., Chapman C., Gloger I. S., Murphy K. E., Southan C. D., Ryan D. M., Smith T. S., Simmons D. L., Walsh F. S., Dingwall C., Christie G. (1999) Identification of a novel aspartic protease (Asp 2) as β-secretase. Mol. Cell Neurosci. 14, 419–427 - PubMed
-
- Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) β-Secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

