Celf1 is required for formation of endoderm-derived organs in zebrafish
- PMID: 24005864
- PMCID: PMC3794766
- DOI: 10.3390/ijms140918009
Celf1 is required for formation of endoderm-derived organs in zebrafish
Abstract
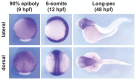
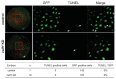
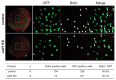
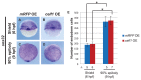
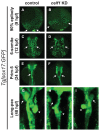
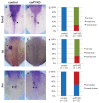
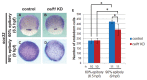
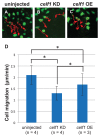
We recently reported that an RNA binding protein called Cugbp Elav-like family member 1 (Celf1) regulates somite symmetry and left-right patterning in zebrafish. In this report, we show additional roles of Celf1 in zebrafish organogenesis. When celf1 is knocked down by using an antisense morpholino oligonucleotides (MO), liver buds fail to form, and pancreas buds do not form a cluster, suggesting earlier defects in endoderm organogenesis. As expected, we found failures in endoderm cell growth and migration during gastrulation in embryos injected with celf1-MOs. RNA immunoprecipitation revealed that Celf1 binds to gata5 and cdc42 mRNAs which are known to be involved in cell growth and migration, respectively. Our results therefore suggest that Celf1 regulates proper organogenesis of endoderm-derived tissues by regulating the expression of such targets.
Figures
Similar articles
-
Celf1 regulation of dmrt2a is required for somite symmetry and left-right patterning during zebrafish development.Development. 2012 Oct;139(19):3553-60. doi: 10.1242/dev.077263. Epub 2012 Aug 16. Development. 2012. PMID: 22899848
-
An evolutionarily conserved kernel of gata5, gata6, otx2 and prdm1a operates in the formation of endoderm in zebrafish.Dev Biol. 2011 Sep 15;357(2):541-57. doi: 10.1016/j.ydbio.2011.06.040. Epub 2011 Jul 1. Dev Biol. 2011. PMID: 21756893
-
Eomesodermin is a localized maternal determinant required for endoderm induction in zebrafish.Dev Cell. 2005 Oct;9(4):523-33. doi: 10.1016/j.devcel.2005.08.010. Dev Cell. 2005. PMID: 16198294
-
The Gata5 target, TGIF2, defines the pancreatic region by modulating BMP signals within the endoderm.Development. 2008 Feb;135(3):451-61. doi: 10.1242/dev.008458. Epub 2007 Dec 19. Development. 2008. PMID: 18094028
-
Multiple roles for Gata5 in zebrafish endoderm formation.Development. 2001 Jan;128(1):125-35. doi: 10.1242/dev.128.1.125. Development. 2001. PMID: 11092818
Cited by
-
Curriculum vitae of CUG binding protein 1 (CELF1) in homeostasis and diseases: a systematic review.Cell Mol Biol Lett. 2024 Mar 5;29(1):32. doi: 10.1186/s11658-024-00556-y. Cell Mol Biol Lett. 2024. PMID: 38443798 Free PMC article.
-
Rho GTPases Signaling in Zebrafish Development and Disease.Cells. 2020 Dec 8;9(12):2634. doi: 10.3390/cells9122634. Cells. 2020. PMID: 33302361 Free PMC article. Review.
-
CUG-BP, Elav-like family member 1 (CELF1) is required for normal myofibrillogenesis, morphogenesis, and contractile function in the embryonic heart.Dev Dyn. 2016 Aug;245(8):854-73. doi: 10.1002/dvdy.24413. Epub 2016 May 31. Dev Dyn. 2016. PMID: 27144987 Free PMC article.
-
Bruno 1/CELF regulates splicing and cytoskeleton dynamics to ensure correct sarcomere assembly in Drosophila flight muscles.PLoS Biol. 2024 Apr 29;22(4):e3002575. doi: 10.1371/journal.pbio.3002575. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 38683844 Free PMC article.
References
-
- Barreau C., Paillard L., Mereau A., Osborne H.B. Mammalian CELF/Bruno-Like RNA-Binding proteins: Molecular characteristics and biological functions. Biochimie. 2006;88:515–525. - PubMed
-
- Takahashi N., Sasagawa N., Suzuki K., Ishiura S. The CUG-Binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 2000;277:518–523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

